5W46
 
 | |
5GIQ
 
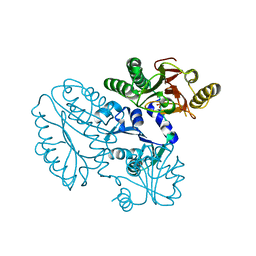 | | Xaa-Pro peptidase from Deinococcus radiodurans, Zinc bound | | 分子名称: | PHOSPHATE ION, Proline dipeptidase, ZINC ION | | 著者 | Are, V.N, Singh, R, Kumar, A, Ghosh, B, Jamdar, S.N, Makde, R.D. | | 登録日 | 2016-06-24 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures and activities of widely conserved small prokaryotic aminopeptidases-P clarify classification of M24B peptidases.
Proteins, 2018
|
|
1XRZ
 
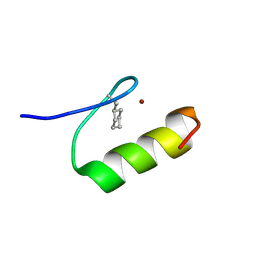 | | NMR Structure of a Zinc Finger with Cyclohexanylalanine Substituted for the Central Aromatic Residue | | 分子名称: | ZINC ION, Zinc finger Y-chromosomal protein | | 著者 | Lachenmann, M.J, Ladbury, J.E, Qian, X, Huang, K, Singh, R, Weiss, M.A. | | 登録日 | 2004-10-17 | | 公開日 | 2004-11-30 | | 最終更新日 | 2021-10-20 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solvation and the hidden thermodynamics of a zinc finger probed
by nonstandard repair of a protein crevice
Protein Sci., 13, 2004
|
|
3QNS
 
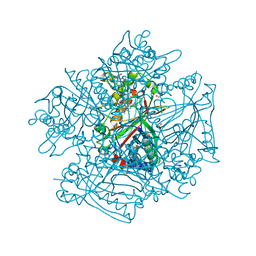 | | DyPB from Rhodococcus jostii RHA1, crystal form 2 | | 分子名称: | DyP Peroxidase, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Grigg, J.C, Roberts, J.N, Singh, R, Eltis, L.D, Murphy, M.E.P. | | 登録日 | 2011-02-09 | | 公開日 | 2011-04-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1.
Biochemistry, 50, 2011
|
|
5GIV
 
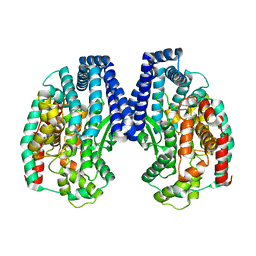 | | Crystal structure of M32 carboxypeptidase from Deinococcus radiodurans R1 | | 分子名称: | ACETATE ION, Carboxypeptidase 1, ZINC ION | | 著者 | Sharma, B, Singh, R, Yadav, P, Ghosh, B, Kumar, A, Jamdar, S.N, Makde, R.D. | | 登録日 | 2016-06-25 | | 公開日 | 2017-07-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Active site gate of M32 carboxypeptidases illuminated by crystal structure and molecular dynamics simulations
Biochim. Biophys. Acta, 1865, 2017
|
|
7DN7
 
 | | Crystal structure of ternary complexes of lactoperoxidase with hydrogen peroxide at 1.70 A resolution | | 分子名称: | 1,2-ETHANEDIOL, 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Singh, P.K, Singh, A.K, Singh, R.P, Kaur, P, Sharma, S, Singh, T.P. | | 登録日 | 2020-12-09 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure of a ternary complex of lactoperoxidase with iodide and hydrogen peroxide at 1.77 angstrom resolution.
J.Inorg.Biochem., 220, 2021
|
|
2KYD
 
 | | RDC and RCSA refinement of an A-form RNA: Improvements in Major Groove Width | | 分子名称: | RNA (5'-R(*CP*UP*AP*GP*UP*UP*AP*GP*CP*UP*AP*AP*CP*UP*AP*G)-3') | | 著者 | Tolbert, B.S, Summers, M.F, Miyazaki, Y, Barton, S, Kinde, B, Stark, P, Singh, R, Bax, A, Case, D. | | 登録日 | 2010-05-24 | | 公開日 | 2010-07-07 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement.
J.Biomol.Nmr, 47, 2010
|
|
4K57
 
 | |
6A4R
 
 | | Crystal structure of aspartate bound peptidase E from Salmonella enterica | | 分子名称: | ASPARTIC ACID, Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-24 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.828 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
6A4S
 
 | | Crystal structure of peptidase E with ordered active site loop from Salmonella enterica | | 分子名称: | Peptidase E | | 著者 | Yadav, P, Chandravanshi, K, Goyal, V.D, Singh, R, Kumar, A, Gokhale, S.M, Makde, R.D. | | 登録日 | 2018-06-20 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of Asp-bound peptidase E from Salmonella enterica: Active site at dimer interface illuminates Asp recognition.
FEBS Lett., 592, 2018
|
|
7FFP
 
 | |
