1HG3
 
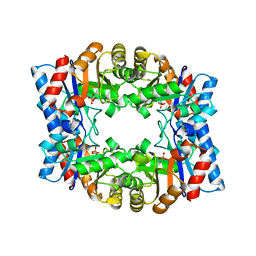 | | Crystal structure of tetrameric TIM from Pyrococcus woesei. | | Descriptor: | 3-PHOSPHONOPROPANOIC ACID, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Walden, H, Bell, G.S, Russell, R.J.M, Siebers, B, Hensel, R, Taylor, G.L. | | Deposit date: | 2000-12-12 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tiny Tim: A Small, Tetrameric, Hyperthermostable Triosephosphate Isomerase
J.Mol.Biol., 306, 2001
|
|
1W8S
 
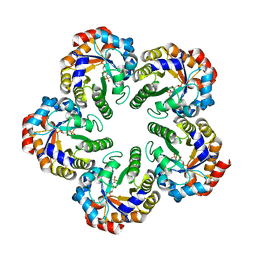 | | The mechanism of the Schiff Base Forming Fructose-1,6-bisphosphate Aldolase: Structural analysis of reaction intermediates | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of the Schiff base forming fructose-1,6-bisphosphate aldolase: structural analysis of reaction intermediates.
Biochemistry, 44, 2005
|
|
2YCE
 
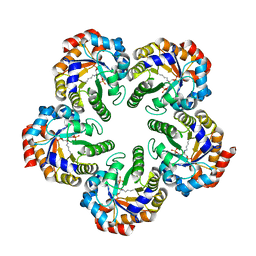 | | Structure of an Archaeal fructose-1,6-bisphosphate aldolase with the catalytic Lys covalently bound to the carbinolamine intermediate of the substrate. | | Descriptor: | D-MANNITOL-1,6-DIPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS 1 | | Authors: | Lorentzen, E, Siebers, B, Hensel, R, Pohl, E. | | Deposit date: | 2011-03-14 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of the Schiff Base Forming Fructose-1,6-Bisphosphate Aldolase: Structural Analysis of Reaction Intermediates.
Biochemistry, 44, 2005
|
|
2R91
 
 | | Crystal Structure of KD(P)GA from T.tenax | | Descriptor: | 2-Keto-3-deoxy-(6-phospho-)gluconate aldolase, SULFATE ION | | Authors: | Pauluhn, A, Pohl, E, Lorentzen, E, Siebers, B, Ahmed, H, Buchinger, S, Schomburg, D. | | Deposit date: | 2007-09-12 | | Release date: | 2008-03-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and stereochemical studies of KD(P)G aldolase from Thermoproteus tenax.
Proteins, 72, 2008
|
|
1OK6
 
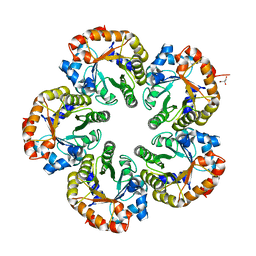 | | Orthorhombic crystal form of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I, GLYCEROL | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-18 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1OJX
 
 | | Crystal structure of an Archaeal fructose 1,6-bisphosphate aldolase | | Descriptor: | FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-16 | | Release date: | 2003-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
1OK4
 
 | | Archaeal fructose 1,6-bisphosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Zwart, P, Stark, A, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2003-07-17 | | Release date: | 2003-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an archaeal class I aldolase and the evolution of (betaalpha)8 barrel proteins.
J. Biol. Chem., 278, 2003
|
|
4CSO
 
 | | The structure of OrfY from Thermoproteus tenax | | Descriptor: | ORFY PROTEIN, TRANSCRIPTION FACTOR | | Authors: | Zeth, K, Hagemann, A, Siebers, B, Martin, J, Lupas, A.N. | | Deposit date: | 2014-03-09 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Challenging the state of the art in protein structure prediction: Highlights of experimental target structures for the 10th Critical Assessment of Techniques for Protein Structure Prediction Experiment CASP10.
Proteins, 82 Suppl 2, 2014
|
|
