7CW2
 
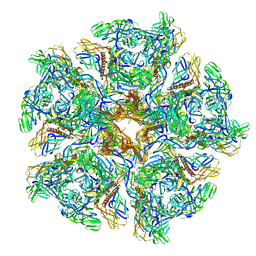 | | Cryo-EM structure of Chikungunya virus in complex with Fab fragments of mAb CHK-263 (subregion around icosahedral 5-fold vertex) | | Descriptor: | Capsid protein, E1 glycoprotein, E2 glycoprotein, ... | | Authors: | Zhou, Q.F, Fox, J.M, Earnest, J.T, Ng, T.S, Kim, A.S, Fibriansah, G, Kostyuchenko, V.A, Shu, B, Diamond, M.S, Lok, S.M. | | Deposit date: | 2020-08-27 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of Chikungunya virus inhibition by monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OP7
 
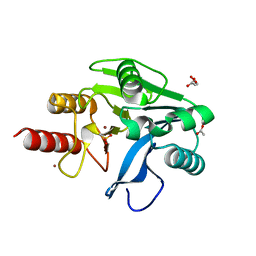 | | Structure of oxidized VIM-20 | | Descriptor: | ACETATE ION, Metallo-beta-lactamase VIM-20, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
6OP6
 
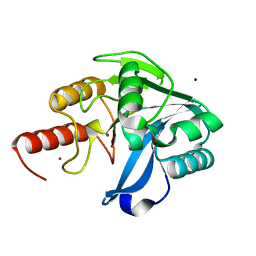 | | Structure of VIM-20 in the reduced state | | Descriptor: | Metallo-beta-lactamase VIM-20, SODIUM ION, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
6T2B
 
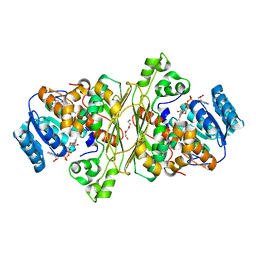 | | Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycosyl hydrolase family 109 protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chaberski, E.K, Fredslund, F, Teze, D, Shuoker, B, Kunstmann, S, Karlsson, E.N, Hachem, M.A, Welner, D.H. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Catalytic Acid-Base in GH109 Resides in a Conserved GGHGG Loop and Allows for Comparable alpha-Retaining and beta-Inverting Activity in an N-Acetylgalactosaminidase from Akkermansia muciniphila
Acs Catalysis, 2020
|
|
3EVR
 
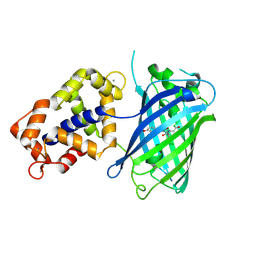 | | Crystal structure of Calcium bound monomeric GCAMP2 | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
3EVP
 
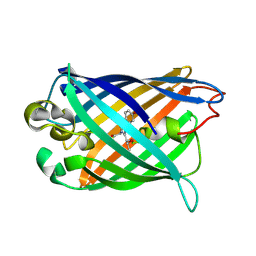 | | crystal structure of circular-permutated EGFP | | Descriptor: | Green fluorescent protein,Green fluorescent protein | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.453 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
3EVU
 
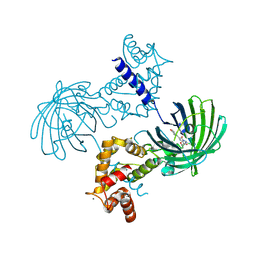 | | Crystal structure of Calcium bound dimeric GCAMP2 | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
3EVV
 
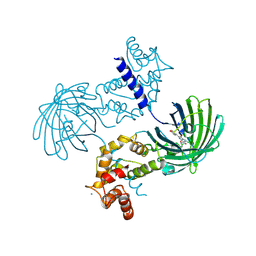 | | Crystal Structure of Calcium bound dimeric GCAMP2 (#2) | | Descriptor: | CALCIUM ION, Myosin light chain kinase, Green fluorescent protein, ... | | Authors: | Wang, Q, Shui, B, Kotlikoff, M.I, Sondermann, H. | | Deposit date: | 2008-10-13 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Calcium Sensing by GCaMP2.
Structure, 16, 2008
|
|
1JVK
 
 | | THREE-DIMENSIONAL STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN DIMER ACTING AS A LETHAL AMYLOID PRECURSOR | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Bourne, P.C, Ramsland, P.A, Shan, L, Fan, Z.-C, DeWitt, C.R, Shultz, B.B, Terzyan, S.S, Edmundson, A.B. | | Deposit date: | 2001-08-30 | | Release date: | 2002-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Three-dimensional structure of an immunoglobulin light-chain dimer with amyloidogenic properties.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
5CGA
 
 | | Structure of Hydroxyethylthiazole kinase ThiM from Staphylococcus aureus in complex with substrate analog 2-(1,3,5-trimethyl-1H-pyrazole-4-yl)ethanol | | Descriptor: | 2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethanol, Hydroxyethylthiazole kinase, MAGNESIUM ION | | Authors: | Kuenz, M, Drebes, J, Windshuegel, B, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
5CGE
 
 | | Structure of Hydroxyethylthiazole Kinase ThiM from Staphylococcus aureus in complex with substrate analog 2-(2-methyl-1H-imidazole-1-yl)ethanol | | Descriptor: | 2-(2-methyl-1H-imidazol-1-yl)ethanol, Hydroxyethylthiazole kinase, MAGNESIUM ION | | Authors: | Kuenz, M, Drebes, J, Windshuegel, B, Cang, H, Wrenger, C, Betzel, C. | | Deposit date: | 2015-07-09 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure of ThiM from Vitamin B1 biosynthetic pathway of Staphylococcus aureus - Insights into a novel pro-drug approach addressing MRSA infections.
Sci Rep, 6, 2016
|
|
