5ZLG
 
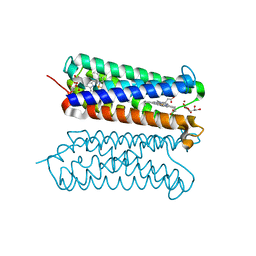 | | Human duodenal cytochrome b (Dcytb) in zinc ion and ascorbate bound form | | Descriptor: | ASCORBIC ACID, Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
5ZLE
 
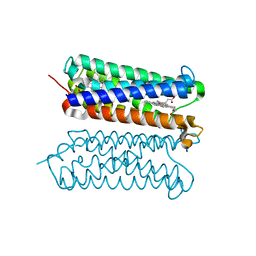 | | Human duodenal cytochrome b (Dcytb) in substrate free form | | Descriptor: | Cytochrome b reductase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ganasen, M, Togashi, H, Mauk, G.A, Shiro, Y, Sawai, H, Sugimoto, H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for promotion of duodenal iron absorption by enteric ferric reductase with ascorbate.
Commun Biol, 1, 2018
|
|
7W79
 
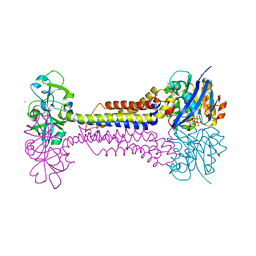 | | Heme exporter HrtBA in complex with Mn-AMPPNP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, Putative ABC transport system integral membrane protein, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7B
 
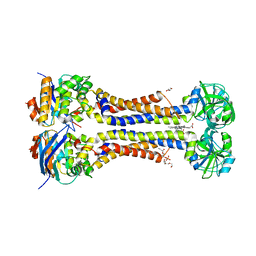 | | Heme exporter HrtBA in complex with protoporphyrin IX containing manganese(III), high resolution data | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING MN, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7A
 
 | | Heme exporter in complex with Mn-containing protoporphyrin IX, Mn-anomalous data | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7D
 
 | | Heme exporter HrtBA in complex with heme | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative ABC transport system integral membrane protein, Putative ABC transport system, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shiro, Y, Shirouzu, M. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W78
 
 | | Heme exporter HrtBA in complex with Mg-AMPPNP | | Descriptor: | ACETATE ION, DODECANE, GLYCEROL, ... | | Authors: | Hisano, T, Nakamura, H, Rahman, M.M, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7W7C
 
 | | Heme exporter in the unliganded form | | Descriptor: | Putative ABC transport system integral membrane protein, Putative ABC transport system, ATP-binding protein, ... | | Authors: | Rahman, M.M, Hisano, T, Nakamura, H, Tosha, T, Shirouzu, M, Shiro, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2Z3U
 
 | | Crystal Structure of Chromopyrrolic Acid Bound Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DI-1H-INDOL-3-YL-1H-PYRROLE-2,5-DICARBOXYLIC ACID, Cytochrome P450, ... | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3A16
 
 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Propionaldoxime | | Descriptor: | (1Z)-propanal oxime, Aldoxime dehydratase, MAGNESIUM ION, ... | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A18
 
 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (soaked crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A17
 
 | | Crystal Structure of Aldoxime Dehydratase (OxdRE) in Complex with Butyraldoxime (Co-crystal) | | Descriptor: | (1Z)-butanal oxime, Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3A15
 
 | | Crystal Structure of Substrate-Free Form of Aldoxime Dehydratase (OxdRE) | | Descriptor: | Aldoxime dehydratase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Kato, Y, Asano, Y, Shiro, Y, Aono, S. | | Deposit date: | 2009-03-26 | | Release date: | 2009-09-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | X-ray crystal structure of michaelis complex of aldoxime dehydratase
J.Biol.Chem., 284, 2009
|
|
3AYF
 
 | | Crystal structure of nitric oxide reductase | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Matsumoto, Y, Tosha, T, Pisliakov, A.V, Hino, T, Sugimoti, H, Nagano, S, Sugita, Y, Shiro, Y. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AYG
 
 | | Crystal structure of nitric oxide reductase complex with HQNO | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, CALCIUM ION, ... | | Authors: | Matsumoto, Y, Tosha, T, Pisliakov, A.V, Hino, T, Sugimoti, H, Nagano, S, Sugita, Y, Shiro, Y. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3VOX
 
 | |
3VOK
 
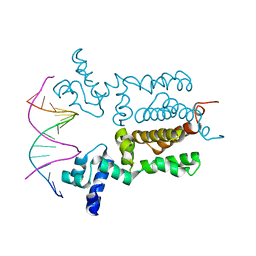 | | X-ray Crystal Structure of Wild Type HrtR in the Apo Form with the Target DNA. | | Descriptor: | 5'-D(*AP*TP*GP*AP*CP*AP*CP*TP*GP*TP*GP*TP*CP*AP*T)-3', Transcriptional regulator | | Authors: | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Transcriptional Regulation of Heme Homeostasis in Lactococcus lactis.
J.Biol.Chem., 287, 2012
|
|
3VP5
 
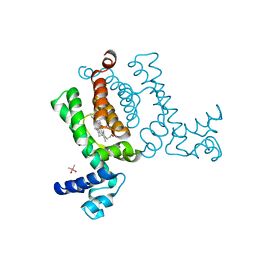 | | X-ray Crystal Structure of Wild Type HrtR in the Holo Form | | Descriptor: | CACODYLATE ION, PROTOPORPHYRIN IX CONTAINING FE, Transcriptional regulator | | Authors: | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | Deposit date: | 2012-02-23 | | Release date: | 2012-07-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Transcriptional Regulation of Heme Homeostasis in Lactococcus lactis.
J.Biol.Chem., 287, 2012
|
|
3WVS
 
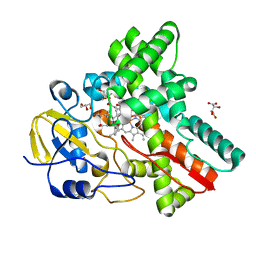 | | Crystal Structure of Cytochrome P450revI | | Descriptor: | (2E,4S,5S,6E,8E)-10-{(2R,3S,6S,8R,9S)-9-butyl-8-[(1E,3E)-4-carboxy-3-methylbuta-1,3-dien-1-yl]-3-methyl-1,7-dioxaspiro[5.5]undec-2-yl}-5-hydroxy-4,8-dimethyldeca-2,6,8-trienoic acid, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Nagano, S, Takahashi, S, Osada, H, Shiro, Y. | | Deposit date: | 2014-06-06 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-function analyses of cytochrome P450revI involved in reveromycin A biosynthesis and evaluation of the biological activity of its substrate, reveromycin T.
J.Biol.Chem., 289, 2014
|
|
3A0R
 
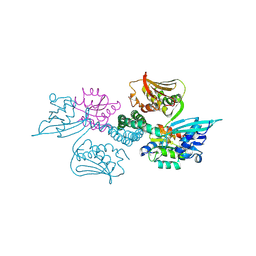 | | Crystal structure of histidine kinase ThkA (TM1359) in complex with response regulator protein TrrA (TM1360) | | Descriptor: | MERCURY (II) ION, Response regulator, Sensor protein | | Authors: | Yamada, S, Sugimoto, H, Kobayashi, M, Ohno, A, Nakamura, H, Shiro, Y. | | Deposit date: | 2009-03-24 | | Release date: | 2009-10-20 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure of PAS-linked histidine kinase and the response regulator complex
Structure, 17, 2009
|
|
3A1L
 
 | | Crystal Structure of 11,11'-Dichlorochromopyrrolic Acid Bound Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 3,4-bis(7-chloro-1H-indol-3-yl)-1H-pyrrole-2,5-dicarboxylic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Theoretical and experimental studies of the conversion of chromopyrrolic acid to an antitumor derivative by cytochrome P450 StaP: the catalytic role of water molecules
J.Am.Chem.Soc., 131, 2009
|
|
2ZQX
 
 | | Cytochrome P450BSbeta cocrystallized with heptanoic acid | | Descriptor: | Cytochrome P450 152A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shoji, O, Fujishiro, T, Nagano, S, Hirose, T, Shiro, Y, Watanabe, Y. | | Deposit date: | 2008-08-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Understanding substrate misrecognition of hydrogen peroxide dependent cytochrome P450 from Bacillus subtilis.
J.Biol.Inorg.Chem., 2010
|
|
2Z6B
 
 | | Crystal Structure Analysis of (gp27-gp5)3 conjugated with Fe(III) protoporphyrin | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, Baseplate structural protein Gp27, Tail-associated lysozyme | | Authors: | Koshiyama, T, Yokoi, N, Ueno, T, Kanamaru, S, Nagano, S, Shiro, Y, Arisaka, F, Watanabe, Y. | | Deposit date: | 2007-07-28 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Molecular design of heteroprotein assemblies providing a bionanocup as a chemical reactor.
Small, 4, 2008
|
|
2ZQJ
 
 | | Substrate-Free Form of Cytochrome P450BSbeta | | Descriptor: | Cytochrome P450 152A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shoji, O, Fujishiro, T, Nagano, S, Hirose, T, Shiro, Y, Watanabe, Y. | | Deposit date: | 2008-08-11 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding substrate misrecognition of hydrogen peroxide dependent cytochrome P450 from Bacillus subtilis.
J.Biol.Inorg.Chem., 2010
|
|
3AWM
 
 | | Cytochrome P450SP alpha (CYP152B1) wild-type with palmitic acid | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Fatty acid alpha-hydroxylase, PALMITIC ACID, ... | | Authors: | Fujishiro, T, Shoji, O, Nagano, S, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2011-03-25 | | Release date: | 2011-06-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of H2O2-dependent cytochrome P450SPalpha with its bound fatty acid substrate: insight into the regioselective hydroxylation of fatty acids at the alpha position
J.Biol.Chem., 286, 2011
|
|
