4IIB
 
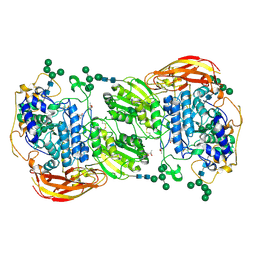 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | 登録日 | 2012-12-20 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIH
 
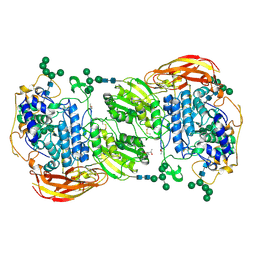 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with thiocellobiose | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | 登録日 | 2012-12-20 | | 公開日 | 2013-04-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IID
 
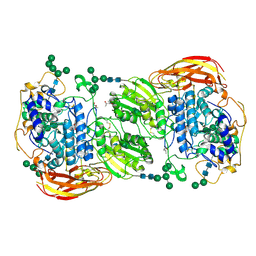 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with 1-deoxynojirimycin | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-DEOXYNOJIRIMYCIN, ... | | 著者 | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | 登録日 | 2012-12-20 | | 公開日 | 2013-04-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
4IIG
 
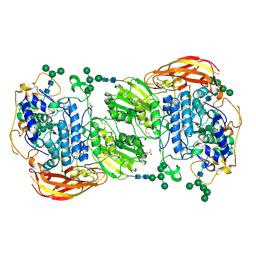 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with D-glucose | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | 登録日 | 2012-12-20 | | 公開日 | 2013-04-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
6KPM
 
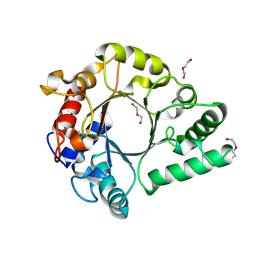 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | 分子名称: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | 著者 | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | 登録日 | 2019-08-15 | | 公開日 | 2019-10-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
4IIE
 
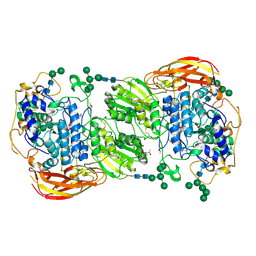 | | Crystal structure of beta-glucosidase 1 from Aspergillus aculeatus in complex with calystegine B(2) | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Suzuki, K, Sumitani, J, Kawaguchi, T, Fushinobu, S. | | 登録日 | 2012-12-20 | | 公開日 | 2013-04-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structures of glycoside hydrolase family 3 beta-glucosidase 1 from Aspergillus aculeatus
Biochem.J., 452, 2013
|
|
2D44
 
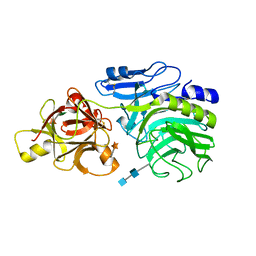 | | Crystal structure of arabinofuranosidase complexed with arabinofuranosyl-alpha-1,2-xylobiose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose-(1-2)-alpha-D-xylopyranose-(1-4)-alpha-D-xylopyranose, alpha-L-arabinofuranosidase B | | 著者 | Miyanaga, A, Koseki, T, Miwa, Y, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | 登録日 | 2005-10-07 | | 公開日 | 2006-09-19 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The family 42 carbohydrate-binding module of family 54 alpha-L-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose
Biochem.J., 399, 2006
|
|
2D43
 
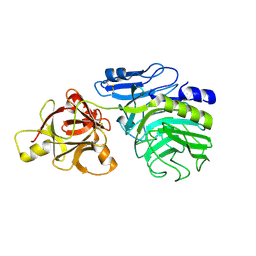 | | Crystal structure of arabinofuranosidase complexed with arabinotriose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-L-arabinofuranose-(1-5)-alpha-L-arabinofuranose, alpha-L-arabinofuranosidase B | | 著者 | Miyanaga, A, Koseki, T, Miwa, Y, Matsuzawa, H, Wakagi, T, Shoun, H, Fushinobu, S. | | 登録日 | 2005-10-07 | | 公開日 | 2006-09-19 | | 最終更新日 | 2021-11-10 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | The family 42 carbohydrate-binding module of family 54 alpha-L-arabinofuranosidase specifically binds the arabinofuranose side chain of hemicellulose
Biochem.J., 399, 2006
|
|
2D0D
 
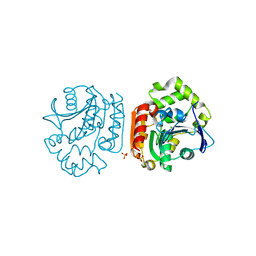 | | Crystal Structure of a Meta-cleavage Product Hydrolase (CumD) A129V Mutant | | 分子名称: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, CHLORIDE ION, PHOSPHATE ION | | 著者 | Jun, S.Y, Fushinobu, S, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | 登録日 | 2005-08-01 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Improving the catalytic efficiency of a meta-cleavage product hydrolase (CumD) from Pseudomonas fluorescens IP01
Biochim.Biophys.Acta, 1764, 2006
|
|
3AFJ
 
 | |
3AHF
 
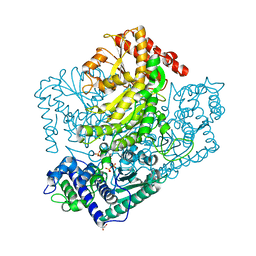 | | Phosphoketolase from Bifidobacterium Breve complexed with inorganic phosphate | | 分子名称: | GLYCEROL, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | 登録日 | 2010-04-22 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
3AHH
 
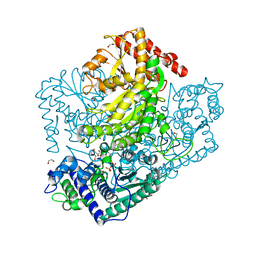 | | H142A mutant of Phosphoketolase from Bifidobacterium Breve complexed with acetyl thiamine diphosphate | | 分子名称: | 1,2-ETHANEDIOL, 2-ACETYL-THIAMINE DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Suzuki, R, Katayama, T, Kim, B.-J, Wakagi, T, Shoun, H, Ashida, H, Yamamoto, K, Fushinobu, S. | | 登録日 | 2010-04-22 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structures of phosphoketolase: thiamine diphosphate-dependent dehydration mechanism
J.Biol.Chem., 285, 2010
|
|
1J1I
 
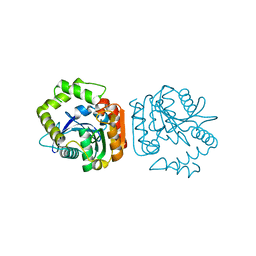 | | Crystal structure of a His-tagged Serine Hydrolase Involved in the Carbazole Degradation (CarC enzyme) | | 分子名称: | meta cleavage compound hydrolase | | 著者 | Habe, H, Morii, K, Fushinobu, S, Nam, J.W, Ayabe, Y, Yoshida, T, Wakagi, T, Yamane, H, Nojiri, H, Omori, T. | | 登録日 | 2002-12-05 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal structure of a histidine-tagged serine hydrolase involved in the carbazole degradation (CarC enzyme).
Biochem.Biophys.Res.Commun., 303, 2003
|
|
7CL7
 
 | |
7CL9
 
 | |
4ZLG
 
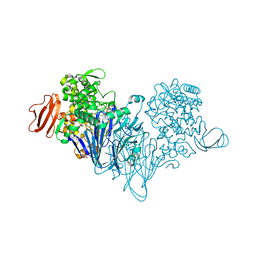 | | Cellobionic acid phosphorylase - gluconic acid complex | | 分子名称: | CHLORIDE ION, D-gluconic acid, D-glucono-1,5-lactone, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZOH
 
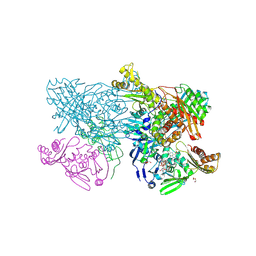 | | Crystal structure of glyceraldehyde oxidoreductase | | 分子名称: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Nishimasu, H, Fushinobu, S, Wakagi, T. | | 登録日 | 2015-05-06 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Archaeal Mo-Containing Glyceraldehyde Oxidoreductase Isozymes Exhibit Diverse Substrate Specificities through Unique Subunit Assemblies.
Plos One, 11, 2016
|
|
4ZLI
 
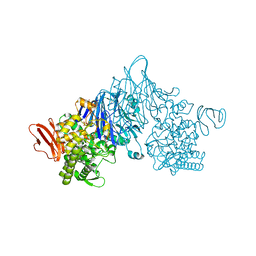 | | Cellobionic acid phosphorylase - 3-O-beta-D-glucopyranosyl-alpha-D-glucopyranuronic acid complex | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLE
 
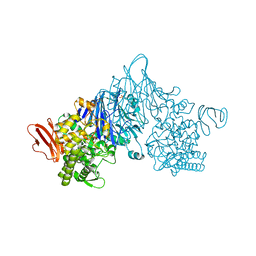 | | Cellobionic acid phosphorylase - ligand free structure | | 分子名称: | CHLORIDE ION, GLYCEROL, Putative b-glycan phosphorylase, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
4ZLF
 
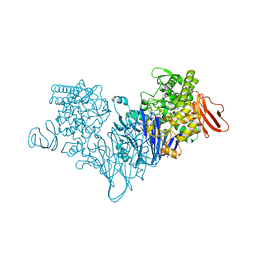 | | Cellobionic acid phosphorylase - cellobionic acid complex | | 分子名称: | 4-O-beta-D-glucopyranosyl-D-gluconic acid, CHLORIDE ION, GLYCEROL, ... | | 著者 | Nam, Y.W, Arakawa, T, Fushinobu, S. | | 登録日 | 2015-05-01 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure and Substrate Recognition of Cellobionic Acid Phosphorylase, Which Plays a Key Role in Oxidative Cellulose Degradation by Microbes.
J.Biol.Chem., 290, 2015
|
|
6KQT
 
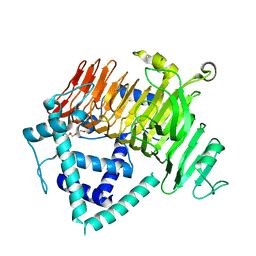 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | 分子名称: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | 登録日 | 2019-08-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
6KQS
 
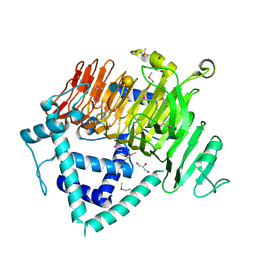 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative | | 分子名称: | GLYCEROL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, lacto-N-biosidase | | 著者 | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | 登録日 | 2019-08-18 | | 公開日 | 2020-06-10 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
3ABB
 
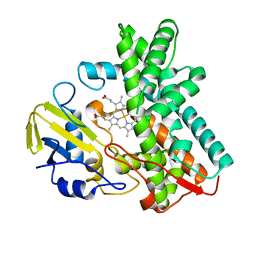 | | Crystal structure of CYP105D6 | | 分子名称: | Cytochrome P450 hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Xu, L.H, Fushinobu, S, Takamatsu, S, Wakagi, T, Ikeda, H, Shoun, H. | | 登録日 | 2009-12-04 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis
J.Biol.Chem., 285, 2010
|
|
3ABA
 
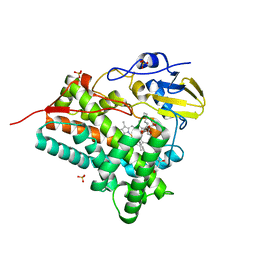 | | Crystal structure of CYP105P1 in complex with filipin I | | 分子名称: | (3R,4S,6S,8S,10R,12R,14R,16S,17E,19E,21E,23E,25E,28R)-3-hexyl-4,6,8,10,12,14,16-heptahydroxy-17,28-dimethyloxacyclooctacosa-17,19,21,23,25-pentaen-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE, ... | | 著者 | Xu, L.H, Fushinobu, S, Takamatsu, S, Wakagi, T, Ikeda, H, Shoun, H. | | 登録日 | 2009-12-04 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis
J.Biol.Chem., 285, 2010
|
|
7V6M
 
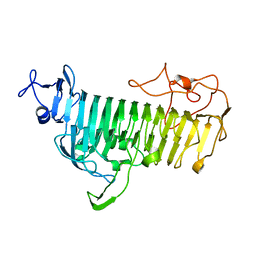 | |
