1QLG
 
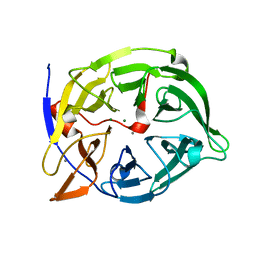 | | Crystal structure of phytase with magnesium from Bacillus amyloliquefaciens | | 分子名称: | 3-PHYTASE, CALCIUM ION, MAGNESIUM ION | | 著者 | Shin, S, Ha, N.-C, Oh, B.-H. | | 登録日 | 1999-08-31 | | 公開日 | 2000-02-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structures of a Novel, Thermostable Phytase in Partially and Fully Calcium-Loaded States
Nat.Struct.Biol., 7, 2000
|
|
7D24
 
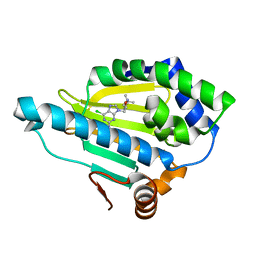 | | Hsp90 alpha N-terminal domain in complex with a 4B compund | | 分子名称: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | 著者 | Shin, S.C, Kim, E.E. | | 登録日 | 2020-09-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D26
 
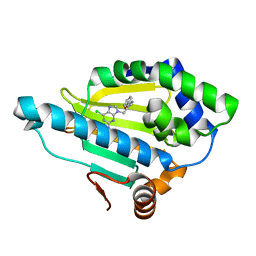 | | Hsp90 alpha N-terminal domain in complex with a 8 compund | | 分子名称: | 6-chloranyl-9-[(2-phenyl-1,3-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | 著者 | Shin, S.C, Kim, E.E. | | 登録日 | 2020-09-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D22
 
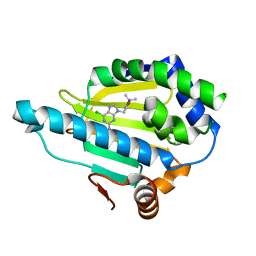 | | Hsp90 alpha N-terminal domain in complex with a 6B compund | | 分子名称: | 9-[(3-tert-butyl-1,2-oxazol-5-yl)methyl]-6-chloranyl-purin-2-amine, Heat shock protein HSP 90-alpha | | 著者 | Shin, S.C, Kim, E.E. | | 登録日 | 2020-09-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D1V
 
 | | Hsp90 alpha N-terminal domain in complex with a 6C compund | | 分子名称: | 6-chloranyl-9-[(3-propan-2-yl-1,2-oxazol-5-yl)methyl]purin-2-amine, Heat shock protein HSP 90-alpha | | 著者 | Shin, S.C, Kim, E.E. | | 登録日 | 2020-09-15 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.332 Å) | | 主引用文献 | Structural Basis for Design of New Purine-Based Inhibitors Targeting the Hydrophobic Binding Pocket of Hsp90.
Int J Mol Sci, 21, 2020
|
|
7D25
 
 | |
1OCH
 
 | |
1GS3
 
 | |
1OBK
 
 | |
1OCM
 
 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COVALENTLY COMPLEXED WITH PYROPHOSPHATE FROM BRADYRHIZOBIUM JAPONICUM | | 分子名称: | MALONAMIDASE E2, PYROPHOSPHATE 2- | | 著者 | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | 登録日 | 2003-02-08 | | 公開日 | 2003-02-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1O9N
 
 | |
1OBI
 
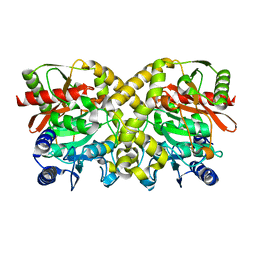 | |
1O9O
 
 | |
1O9P
 
 | |
1OCL
 
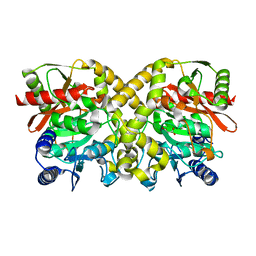 | | THE CRYSTAL STRUCTURE OF MALONAMIDASE E2 COMPLEXED WITH MALONATE FROM BRADYRHIZOBIUM JAPONICUM | | 分子名称: | MALONAMIDASE E2, MALONIC ACID | | 著者 | Shin, S, Ha, N.-C, Lee, T.-H, Oh, B.-H. | | 登録日 | 2003-02-08 | | 公開日 | 2003-02-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Characterization of a Novel Ser-Cisser-Lys Catalytic Triad in Comparison with the Classical Ser-His-Asp Triad.
J.Biol.Chem., 278, 2003
|
|
1OBL
 
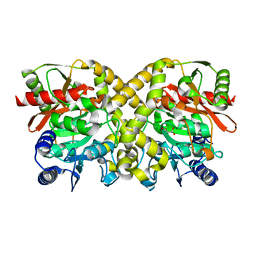 | |
1O9Q
 
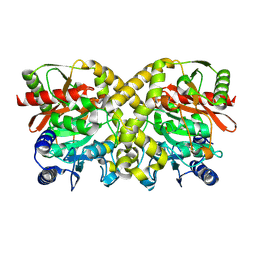 | |
1OCK
 
 | |
1OBJ
 
 | |
1CVM
 
 | | CADMIUM INHIBITED CRYSTAL STRUCTURE OF PHYTASE FROM BACILLUS AMYLOLIQUEFACIENS | | 分子名称: | CADMIUM ION, CALCIUM ION, PHYTASE | | 著者 | Shin, S, Ha, N.-C, Oh, B.-H. | | 登録日 | 1999-08-24 | | 公開日 | 2000-02-24 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
1H6L
 
 | |
1BK1
 
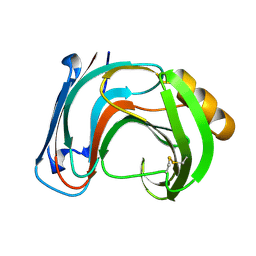 | | ENDO-1,4-BETA-XYLANASE C | | 分子名称: | ENDO-1,4-B-XYLANASE C | | 著者 | Fushinobu, S, Ito, K, Konno, M, Wakagi, T, Matsuzawa, H. | | 登録日 | 1998-07-14 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystallographic and mutational analyses of an extremely acidophilic and acid-stable xylanase: biased distribution of acidic residues and importance of Asp37 for catalysis at low pH.
Protein Eng., 11, 1998
|
|
8HVC
 
 | |
8HVD
 
 | |
4ZXE
 
 | | X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. | | 分子名称: | 1,2-ETHANEDIOL, Glucanase/Chitosanase, SULFATE ION | | 著者 | Shinya, S, Oi, H, Kitaoku, Y, Ohnuma, T, Numata, T, Fukamizo, T. | | 登録日 | 2015-05-20 | | 公開日 | 2016-04-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Mechanism of chitosan recognition by CBM32 carbohydrate-binding modules from a Paenibacillus sp. IK-5 chitosanase/glucanase
Biochem.J., 473, 2016
|
|
