5H41
 
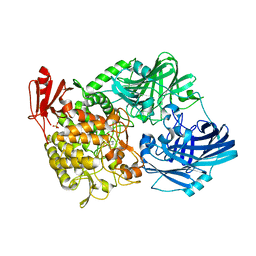 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans in complex with sophorose, isofagomine, sulfate ion | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, SULFATE ION, Uncharacterized protein, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
3X2N
 
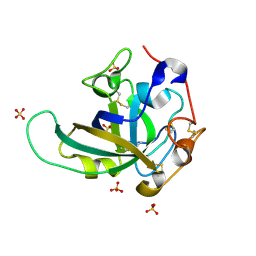 | | Proton relay pathway in inverting cellulase | | Descriptor: | Endoglucanase V-like protein, SULFATE ION | | Authors: | Nakamura, A, Ishida, T, Fushinobu, S, Igarashi, K, Samejima, M. | | Deposit date: | 2014-12-22 | | Release date: | 2015-10-14 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | "Newton's cradle" proton relay with amide-imidic acid tautomerization in inverting cellulase visualized by neutron crystallography.
Sci Adv, 1, 2015
|
|
6KPO
 
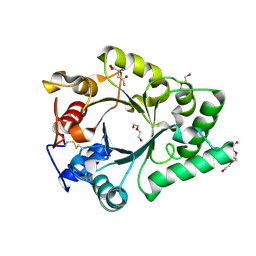 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine-Asn | | Descriptor: | ASPARAGINE, Chitinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPN
 
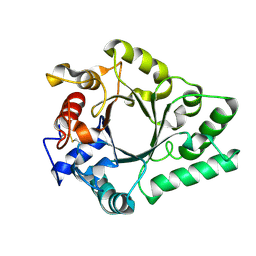 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris D154N/E156Q mutant in complex with fucosyl-N-acetylglucosamine | | Descriptor: | Chitinase, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
5H3Z
 
 | | Crystal Structure of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Nakajima, M, Tanaka, N, Furukawa, N, Nihira, T, Kodutsumi, Y, Takahashi, Y, Sugimoto, N, Miyanaga, A, Fushinobu, S, Taguchi, H, Nakai, H. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-01 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic insight into the substrate specificity of 1,2-beta-oligoglucan phosphorylase from Lachnoclostridium phytofermentans
Sci Rep, 7, 2017
|
|
6KPM
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in complex with L-fucose | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6KPL
 
 | | Crystal Structure of endo-beta-N-acetylglucosaminidase from Cordyceps militaris in apo form | | Descriptor: | Chitinase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Seki, H, Arakawa, T, Yamada, C, Takegawa, K, Fushinobu, S. | | Deposit date: | 2019-08-15 | | Release date: | 2019-10-02 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the specific cleavage of core-fucosylatedN-glycans by endo-beta-N-acetylglucosaminidase from the fungusCordyceps militaris.
J.Biol.Chem., 294, 2019
|
|
6M3X
 
 | | Cryo-EM structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | Descriptor: | FE (III) ION, Sulfur oxygenase/reductase | | Authors: | Sato, Y, Adachi, N, Moriya, T, Arakawa, T, Kawasaki, M, Yamada, C, Senda, T, Fushinobu, S. | | Deposit date: | 2020-03-04 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
6M35
 
 | | Crystal structure of sulfur oxygenase reductase from Sulfurisphaera tokodaii | | Descriptor: | FE (III) ION, GLYCEROL, SULFATE ION, ... | | Authors: | Sato, Y, Yabuki, T, Arakawa, T, Yamada, C, Fushinobu, S, Wakagi, T. | | Deposit date: | 2020-03-02 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallographic and cryogenic electron microscopic structures and enzymatic characterization of sulfur oxygenase reductase fromSulfurisphaera tokodaii.
J Struct Biol X, 4, 2020
|
|
7YQS
 
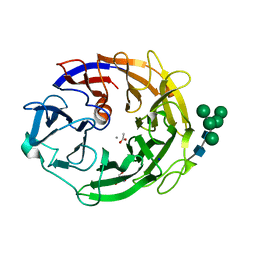 | | Neutron structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, L-Rha complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Yano, N, Kondo, T, Kusaka, K, Yamada, T, Arakawa, T, Sakamoto, T, Fushinobu, S. | | Deposit date: | 2022-08-08 | | Release date: | 2023-08-09 | | Last modified: | 2024-03-27 | | Method: | NEUTRON DIFFRACTION (1.25 Å), X-RAY DIFFRACTION | | Cite: | Charge neutralization and beta-elimination cleavage mechanism of family 42 L-rhamnose-alpha-1,4-D-glucuronate lyase revealed using neutron crystallography.
J.Biol.Chem., 300, 2024
|
|
6JU1
 
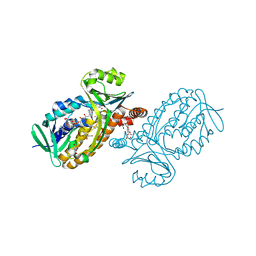 | | p-Hydroxybenzoate hydroxylase Y385F mutant complexed with 3,4-dihydroxybenzoate | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 4-hydroxybenzoate 3-monooxygenase, ... | | Authors: | Yato, M, Arakawa, T, Yamada, C, Fushinobu, S. | | Deposit date: | 2019-04-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Understanding the Molecular Mechanism Underlying the High Catalytic Activity ofp-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid.
Biochemistry, 58, 2019
|
|
2CVQ
 
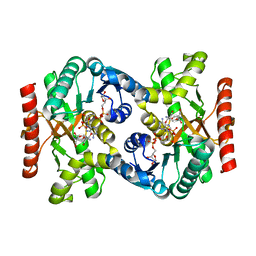 | | Crystal structure of NAD(H)-dependent malate dehydrogenase complexed with NADPH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2005-06-13 | | Release date: | 2005-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structure of NAD-dependent malate dehydrogenase complexed with NADP(H)
Biochem.Biophys.Res.Commun., 334, 2005
|
|
5B1O
 
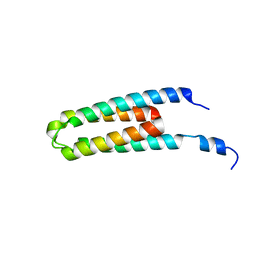 | | DHp domain structure of EnvZ P248A mutant | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
5B1N
 
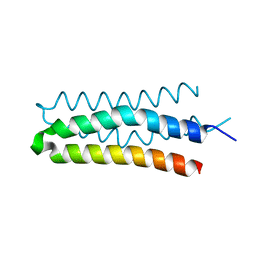 | | DHp domain structure of EnvZ from Escherichia coli | | Descriptor: | Osmolarity sensor protein EnvZ | | Authors: | Okajima, T, Eguchi, Y, Tochio, N, Inukai, Y, Shimizu, R, Ueda, S, Shinya, S, Kigawa, T, Fukamizo, T, Igarashi, M, Utsumi, R. | | Deposit date: | 2015-12-09 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Angucycline antibiotic waldiomycin recognizes common structural motif conserved in bacterial histidine kinases
J. Antibiot., 70, 2017
|
|
5B0R
 
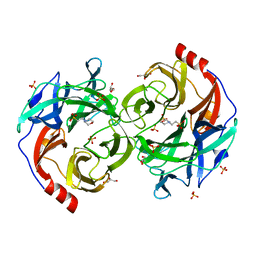 | | Beta-1,2-Mannobiose phosphorylase from Listeria innocua - beta-1,2-mannobiose complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Lin0857 protein, ... | | Authors: | Tsuda, T, Arakawa, T, Fushinobu, S. | | Deposit date: | 2015-11-02 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization and crystal structure determination of beta-1,2-mannobiose phosphorylase from Listeria innocua
Febs Lett., 589, 2015
|
|
6A3J
 
 | | Levoglucosan dehydrogenase, complex with NADH and L-sorbose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative dehydrogenase, ... | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6A3F
 
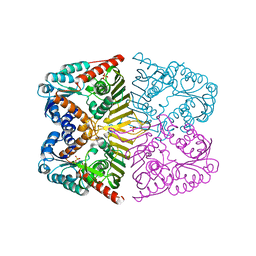 | | Levoglucosan dehydrogenase, apo form | | Descriptor: | Putative dehydrogenase, SULFATE ION | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6KQT
 
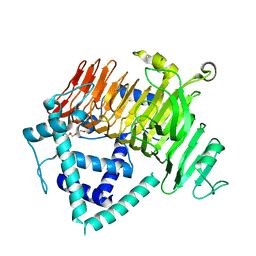 | | Crystal Structure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein | | Descriptor: | SODIUM ION, TRIETHYLENE GLYCOL, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yamada, C, Arakawa, T, Pichler, M.J, Abou Hachem, M, Fushinobu, S. | | Deposit date: | 2019-08-18 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways.
Nat Commun, 11, 2020
|
|
5TD7
 
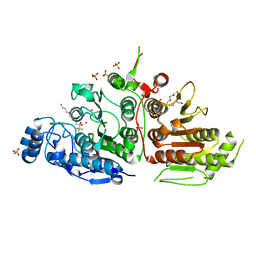 | | Crystal structure of histone deacetylase 10 | | Descriptor: | 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptane-2,2-diol, PENTAETHYLENE GLYCOL, PHOSPHATE ION, ... | | Authors: | Hai, Y, Shinsky, S.A, Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-17 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Histone deacetylase 10 structure and molecular function as a polyamine deacetylase.
Nat Commun, 8, 2017
|
|
5DGQ
 
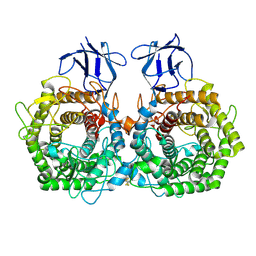 | |
5YHS
 
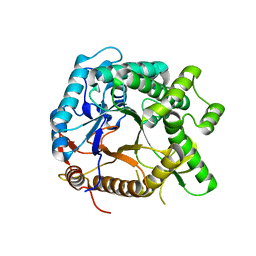 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, apo form | | Descriptor: | Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-09-29 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
5YIF
 
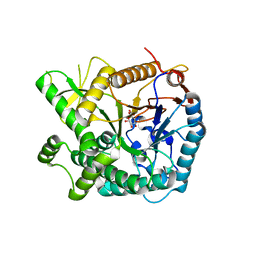 | | Pyruvylated beta-D-galactosidase from Bacillus sp. HMA207, E163A mutant pyruvylated beta-D-galactose complex | | Descriptor: | (2R,4aR,6R,7R,8R,8aR)-2-methyl-6,7,8-tris(oxidanyl)-4,4a,6,7,8,8a-hexahydropyrano[3,2-d][1,3]dioxine-2-carboxylic acid, Pyruvylated beta-D-galactosidase | | Authors: | Tanuma, M, Yamada, C, Arakawa, T, Higuchi, Y, Takegawa, K, Fushinobu, S. | | Deposit date: | 2017-10-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification and characterization of a novel beta-D-galactosidase that releases pyruvylated galactose.
Sci Rep, 8, 2018
|
|
5DGR
 
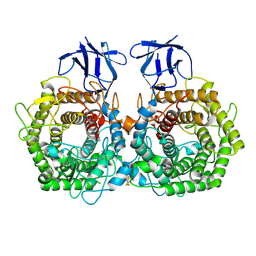 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | Authors: | Suzuki, K, Honda, Y, Fushinobu, S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
7ESN
 
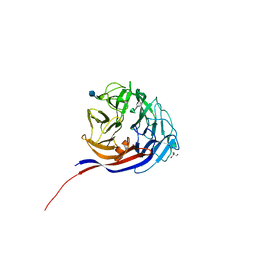 | | Crystal structure of a L-rhamnose-alpha-1,4-D-glucuronate lyase from Fusarium oxysporum 12S, H105F Rha-GlcA complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, L-Rhamnose-alpha-1,4-D-glucuronate lyase, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2021-05-11 | | Release date: | 2021-08-04 | | Last modified: | 2021-09-01 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural and functional analysis of gum arabic l-rhamnose-alpha-1,4-d-glucuronate lyase establishes a novel polysaccharide lyase family.
J.Biol.Chem., 297, 2021
|
|
7DFS
 
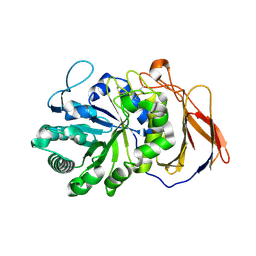 | | Crystal structure of a novel 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase from Fusarium oxysporum 12S - Rha-GlcA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-alpha-L-rhamnosyl-beta-D-glucuronidase, alpha-D-mannopyranose, ... | | Authors: | Kondo, T, Arakawa, T, Fushinobu, S, Sakamoto, T. | | Deposit date: | 2020-11-09 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Biochemical and structural characterization of a novel 4-O-alpha-l-rhamnosyl-beta-d-glucuronidase from Fusarium oxysporum.
Febs J., 288, 2021
|
|
