8I76
 
 | |
8I74
 
 | |
5Y72
 
 | | DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y7C
 
 | | Hapalindole A and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole A | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y84
 
 | | Hapalindole U and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole U | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z43
 
 | | Crystal structure of prenyltransferase AmbP1 apo structure | | Descriptor: | AmbP1, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.361 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z45
 
 | | Crystal structure of prenyltransferase AmbP1 pH6.5 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1BHI
 
 | | STRUCTURE OF TRANSACTIVATION DOMAIN OF CRE-BP1/ATF-2, NMR, 20 STRUCTURES | | Descriptor: | CRE-BP1 | | Authors: | Nagadoi, A, Nakazawa, K, Uda, H, Maekawa, T, Ishii, S, Nishimura, Y. | | Deposit date: | 1998-06-09 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain.
J.Mol.Biol., 287, 1999
|
|
5Z44
 
 | | Crystal structure of prenyltransferase AmbP1 complexed with GSPP | | Descriptor: | AmbP1, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.458 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
4TWW
 
 | | Structure of SARS-3CL protease complex with a Bromobenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(4-bromobenzoyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
4TWY
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
5Z46
 
 | | Crystal structure of prenyltransferase AmbP1 pH8 complexed with GSPP and cis-indolyl vinyl isonitrile | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, AmbP1, GERANYL S-THIOLODIPHOSPHATE, ... | | Authors: | Awakawa, T, Nakashima, Y, Mori, T, Abe, I. | | Deposit date: | 2018-01-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular Insight into the Mg2+-Dependent Allosteric Control of Indole Prenylation by Aromatic Prenyltransferase AmbP1
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6AJV
 
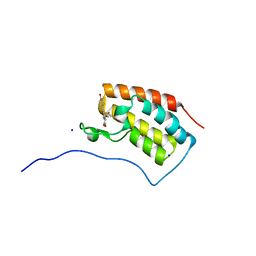 | | Crystal structure of BRD4 in complex with isoliquiritigenin and DMSO (Cocktail No. 3) | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
5Y4G
 
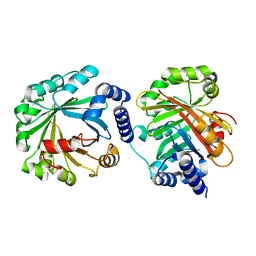 | | Apo Structure of AmbP3 | | Descriptor: | AmbP3 | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-03 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5C5N
 
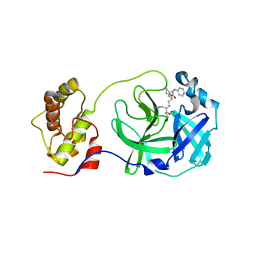 | | Structure of SARS-3CL protease complex with a phenyl-beta-alanyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-3-(1H-imidazol-5-yl)-2-({[(3R,4aS,8aR)-2-(N-phenyl-beta-alanyl)decahydroisoquinolin-3-yl]methyl}amino)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2015-06-21 | | Release date: | 2016-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Fused-ring structure of N-decalin as a novel scaffold for SARS 3CL protease inhibitors
to be published
|
|
3APM
 
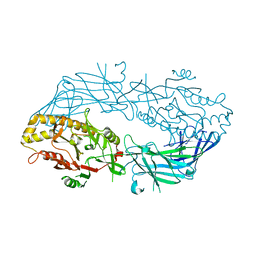 | | Crystal structure of the human SNP PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6AJX
 
 | | Crystal structure of BRD4 in complex with isoliquiritigenin in the absence of DMSO | | Descriptor: | 2',4,4'-TRIHYDROXYCHALCONE, Bromodomain-containing protein 4, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.887 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJY
 
 | | Crystal structure of BRD4 in complex with 2',4'-dihydroxy-2-methoxychalcone | | Descriptor: | 2',4'-dihydroxy-2-methoxychalcone, Bromodomain-containing protein 4, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJZ
 
 | | Joint nentron and X-ray structure of BRD4 in complex with colchicin | | Descriptor: | Bromodomain-containing protein 4, N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide, SODIUM ION | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | NEUTRON DIFFRACTION (1.301 Å), X-RAY DIFFRACTION | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
6AJW
 
 | | Crystal structure of BRD4 in complex with DMSO (Cocktail No. 4) | | Descriptor: | Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, SODIUM ION | | Authors: | Yokoyama, T, Matsumoto, K, Nabeshima, Y, Mizuguchi, M. | | Deposit date: | 2018-08-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Structural and thermodynamic characterization of the binding of isoliquiritigenin to the first bromodomain of BRD4.
Febs J., 286, 2019
|
|
3APN
 
 | | Crystal structure of the human wild-type PAD4 protein | | Descriptor: | Protein-arginine deiminase type-4 | | Authors: | Horikoshi, N, Tachiwana, H, Saito, K, Osakabe, A, Sato, M, Yamada, M, Akashi, S, Nishimura, Y, Kagawa, W, Kurumizaka, H. | | Deposit date: | 2010-10-19 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical analyses of the human PAD4 variant encoded by a functional haplotype gene
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3U2J
 
 | | Neutron crystal structure of human Transthyretin | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
3U2I
 
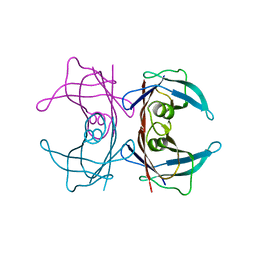 | | X-ray crystal structure of human Transthyretin at room temperature | | Descriptor: | Transthyretin | | Authors: | Yokoyama, T, Mizuguchi, M, Nabeshima, Y, Kusaka, K, Yamada, T, Hosoya, T, Ohhara, T, Kurihara, K, Tomoyori, K, Tanaka, I, Niimura, N. | | Deposit date: | 2011-10-03 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hydrogen-bond network and pH sensitivity in transthyretin: Neutron crystal structure of human transthyretin
J.Struct.Biol., 177, 2012
|
|
7BUL
 
 | |
2RVM
 
 | |
