5B7A
 
 | |
2KQS
 
 | |
5GHC
 
 | |
5GHB
 
 | |
5GHD
 
 | |
2N1W
 
 | |
2N1V
 
 | |
5XF9
 
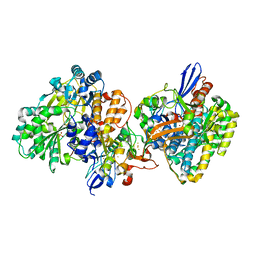 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the air-oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
2RUH
 
 | | Chemical Shift Assignments for MIP and MDM2 in bound state | | Descriptor: | E3 ubiquitin-protein ligase Mdm2 | | Authors: | Nagata, T, Shirakawa, K, Kobayashi, N, Shiheido, H, Horisawa, K, Katahira, M, Doi, N, Yanagawa, H. | | Deposit date: | 2014-06-03 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Inhibition of the MDM2:p53 Interaction by an Optimized MDM2-Binding Peptide Selected with mRNA Display
Plos One, 9, 2014
|
|
5XFA
 
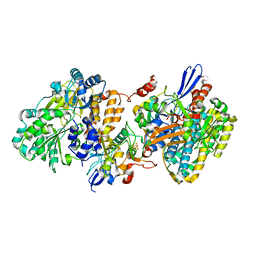 | | Crystal structure of NAD+-reducing [NiFe]-hydrogenase in the H2-reduced state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Shomura, Y, Taketa, M, Nakashima, H, Tai, H, Nakagawa, H, Ikeda, Y, Ishii, M, Igarashi, Y, Nishihara, H, Yoon, K.S, Ogo, S, Hirota, S, Higuchi, Y. | | Deposit date: | 2017-04-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the redox switches in the NAD(+)-reducing soluble [NiFe]-hydrogenase
Science, 357, 2017
|
|
5B3I
 
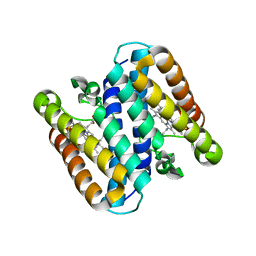 | | Homo-dimeric structure of cytochrome c' from Thermophilic Hydrogenophilus thermoluteolus | | Descriptor: | Cytochrome c prime, HEME C | | Authors: | Fujii, S, Oki, H, Kawahara, K, Yamane, D, Yamanaka, M, Maruno, T, Kobayashi, Y, Masanari, M, Wakai, S, Nishihara, H, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional insights into thermally stable cytochrome c' from a thermophile
Protein Sci., 26, 2017
|
|
5Y34
 
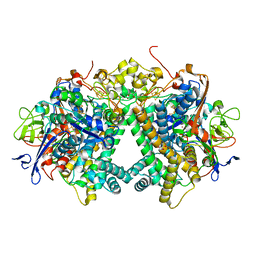 | | Membrane-bound respiratory [NiFe]-hydrogenase from Hydrogenovibrio marinus in a ferricyanide-oxidized condition | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2017-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3WHJ
 
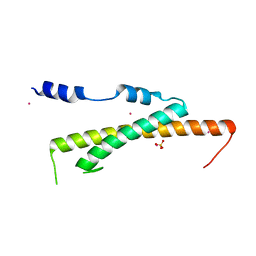 | | Crystal structure of Nas2 N-terminal domain | | Descriptor: | CADMIUM ION, Probable 26S proteasome regulatory subunit p27, SULFATE ION | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHL
 
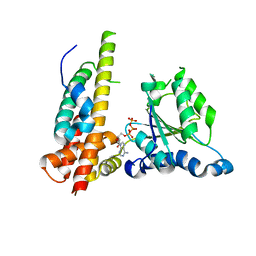 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHK
 
 | | Crystal structure of PAN-Rpt5C chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | Authors: | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | Deposit date: | 2013-08-26 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3AYX
 
 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an H2-reduced condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3AYZ
 
 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an air-oxidized condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
