4YTR
 
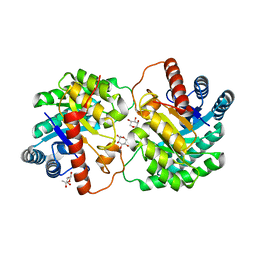 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
4YTS
 
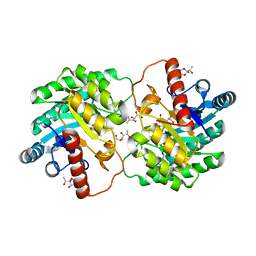 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy 3-keto D-galactitol | | Descriptor: | 1-deoxy-D-xylo-hex-3-ulose, 1-deoxy-alpha-D-xylo-hex-3-ulofuranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
8JQ3
 
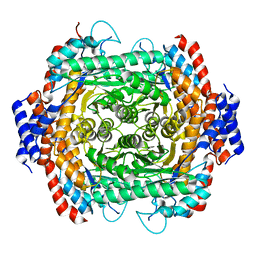 | |
4Q0V
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
7WRR
 
 | |
7WRT
 
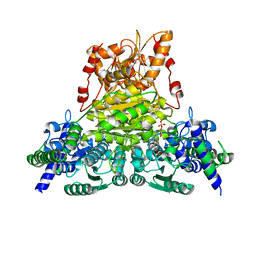 | |
1COO
 
 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
