6WPP
 
 | | HUMAN NIK IN COMPLEX WITH LIGAND COMPOUND X | | Descriptor: | 5-fluoro-N-methyl-2-(7H-pyrrolo[2,3-d]pyrimidin-5-yl)pyrimidin-4-amine, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Shih, A.Y, Hack, M, Mirzadegan, T. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Impact of Protein Preparation on Resulting Accuracy of FEP Calculations.
J.Chem.Inf.Model., 60, 2020
|
|
1WIW
 
 | |
2CVJ
 
 | |
6MRG
 
 | | FAAH bound to non covalent inhibitor | | Descriptor: | (1R)-2-{[6-(2,3-dihydro-1,4-benzodioxin-6-yl)pyrimidin-4-yl]amino}-1-phenylethan-1-ol, Fatty-acid amide hydrolase 1 | | Authors: | Saha, A, Shih, A, Mirzadegan, T, Seierstad, M. | | Deposit date: | 2018-10-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Predicting the Binding of Fatty Acid Amide Hydrolase Inhibitors by Free Energy Perturbation.
J Chem Theory Comput, 14, 2018
|
|
4Q0Q
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0U
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
5J8L
 
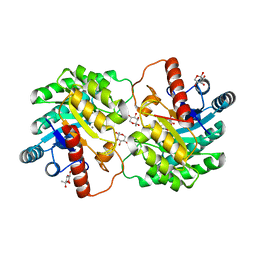 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose, using a crystal grown in microgravity | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Izumori, K, Kamitori, S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
4Q0S
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with ribitol | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), D-ribitol, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0P
 
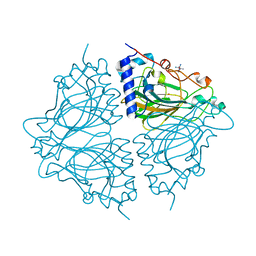 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0V
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
8JQ5
 
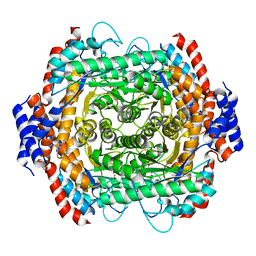 | |
8JQ4
 
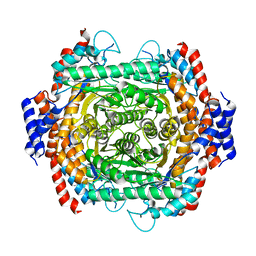 | |
8JQ6
 
 | |
4XSL
 
 | | Crystal strcutre of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with glycerol | | Descriptor: | D-tagatose 3-epimerase, GLYCEROL, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
8JQ3
 
 | |
4YTU
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with L-erythrulose | | Descriptor: | D-tagatose 3-epimerase, L-Erythrulose, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
5X5I
 
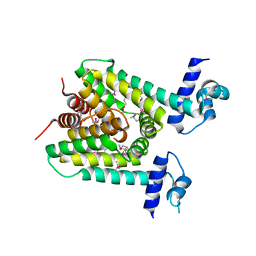 | | The X-ray crystal structure of a TetR family transcription regulator RcdA involved in the regulation of biofilm formation in Escherichia coli | | Descriptor: | HTH-type transcriptional regulator RcdA | | Authors: | Sugino, H, Usui, M, Shimada, T, Nakano, M, Ogasawara, H, Ishihama, A, Hirata, A. | | Deposit date: | 2017-02-16 | | Release date: | 2017-12-27 | | Method: | X-RAY DIFFRACTION (2.554 Å) | | Cite: | A structural sketch of RcdA, a transcription factor controlling the master regulator of biofilm formation.
FEBS Lett., 591, 2017
|
|
4YTR
 
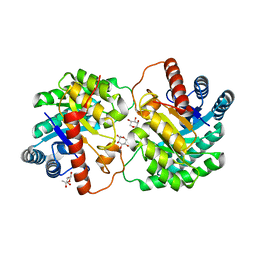 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy L-tagatose | | Descriptor: | 1-deoxy-L-tagatose, 1-deoxy-beta-L-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
4YTQ
 
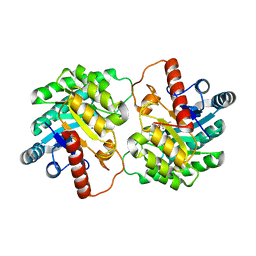 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy D-tagatose | | Descriptor: | 1-deoxy-D-tagatose, 1-deoxy-alpha-D-tagatopyranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
4YTS
 
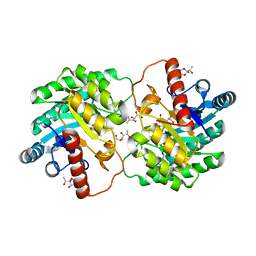 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 1-deoxy 3-keto D-galactitol | | Descriptor: | 1-deoxy-D-xylo-hex-3-ulose, 1-deoxy-alpha-D-xylo-hex-3-ulofuranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
4YTT
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with 6-deoxy L-psicose | | Descriptor: | 6-deoxy-L-psicose, 6-deoxy-alpha-L-psicofuranose, D-tagatose 3-epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-03-18 | | Release date: | 2016-03-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
7CJ4
 
 | |
7CJ9
 
 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. with additional C-terminal residues | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-fructose, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ7
 
 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with L-tagatose | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)oxane-2,3,4,5-tetrol, Epimerase, L-sorbose, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ6
 
 | |
