5Y5F
 
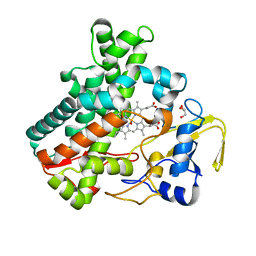 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5M
 
 | | SFX structure of cytochrome P450nor: a complete dark data without pump laser (resting state) | | Descriptor: | NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
4AX7
 
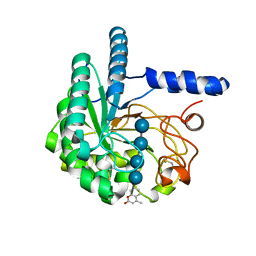 | | Hypocrea jecorina Cel6A D221A mutant soaked with 4-Methylumbelliferyl- beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-06-11 | | Release date: | 2013-01-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4AU0
 
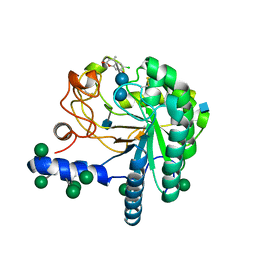 | | Hypocrea jecorina Cel6A D221A mutant soaked with 6-chloro-4- methylumbelliferyl-beta-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-7-hydroxy-4-methyl-2H-chromen-2-one, EXOGLUCANASE 2, ... | | Authors: | Wu, M, Nerinckx, W, Piens, K, Ishida, T, Hansson, H, Stahlberg, J, Sandgren, M. | | Deposit date: | 2012-05-11 | | Release date: | 2013-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design, Synthesis, Evaluation and Enzyme-Substrate Structures of Improved Fluorogenic Substrates for Family 6 Glycoside Hydrolases.
FEBS J., 280, 2013
|
|
4GNA
 
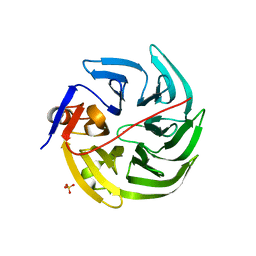 | | mouse SMP30/GNL-xylitol complex | | Descriptor: | CALCIUM ION, Regucalcin, SULFATE ION, ... | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
4GNC
 
 | | human SMP30/GNL-1,5-AG complex | | Descriptor: | 1,5-anhydro-D-glucitol, CALCIUM ION, Regucalcin | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
6K9Z
 
 | | STRUCTURE OF URIDYLYLTRANSFERASE MUTANT | | Descriptor: | ACETATE ION, FE (III) ION, Galactose-1-phosphate uridylyltransferase, ... | | Authors: | Sakuraba, H, Ohshida, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-06-19 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Unique active site formation in a novel galactose 1-phosphate uridylyltransferase from the hyperthermophilic archaeon Pyrobaculum aerophilum.
Proteins, 88, 2020
|
|
7BYT
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A with galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
7BYS
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CITRIC ACID, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
7BYV
 
 | | Crystal structure of exo-beta-1,3-galactanase from Phanerochaete chrysosporium Pc1,3Gal43A E208Q with beta-1,3-galactotriose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Galactan 1,3-beta-galactosidase, ... | | Authors: | Matsuyama, K, Ishida, T, Kishine, N, Fujimoto, Z, Igarashi, K, Kaneko, S. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unique active-site and subsite features in the arabinogalactan-degrading GH43 exo-beta-1,3-galactanase from Phanerochaete chrysosporium .
J.Biol.Chem., 295, 2020
|
|
1O5P
 
 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
6K5Z
 
 | | Structure of uridylyltransferase | | Descriptor: | FE (III) ION, Galactose-1-phosphate uridylyltransferase, PHOSPHATE ION, ... | | Authors: | Sakuraba, H, Ohshida, T, Yoneda, K, Ohshima, T. | | Deposit date: | 2019-05-31 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Unique active site formation in a novel galactose 1-phosphate uridylyltransferase from the hyperthermophilic archaeon Pyrobaculum aerophilum.
Proteins, 88, 2020
|
|
4GN9
 
 | | mouse SMP30/GNL-glucose complex | | Descriptor: | CALCIUM ION, Regucalcin, SULFATE ION, ... | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
7FGL
 
 | | The complex structure of PHF core domain peptide of tau, VQIVYK, and antibody's Fab domain. | | Descriptor: | AMMONIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Tsuchida, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The cross-reaction complex structure with VQIVYK of tau and the antibody's Fab domain.
To Be Published
|
|
7FGK
 
 | | The Fab antibody single structure against tau protein. | | Descriptor: | Fab Heavy Chain, Fab Light Chain, GLYCEROL, ... | | Authors: | Tsuchida, T, Susa, K, Kibiki, T, Tsuchiya, T, Miyamoto, K, In, Y, Minoura, K, Taniguchi, T, Ishida, T, Tomoo, K. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The free structure of the Fab domain of antibody that recognizes the PHF core region VQIINK in Tau protein.
To Be Published
|
|
2E4P
 
 | | Crystal structure of BphA3 (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER, SULFATE ION, ... | | Authors: | Senda, M, Kishigami, S, Kimura, S, Ishida, T, Fukuda, M, Senda, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
2E4Q
 
 | | Crystal structure of BphA3 (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Senda, M, Kishigami, S, Kimura, S, Ishida, T, Fukuda, M, Senda, T. | | Deposit date: | 2006-12-15 | | Release date: | 2007-10-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
1Q3S
 
 | | Crystal structure of the chaperonin from Thermococcus strain KS-1 (FormIII crystal complexed with ADP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Thermosome alpha subunit | | Authors: | Shomura, Y, Yoshida, T, Iizuka, R, Maruyama, T, Yohda, M, Miki, K. | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of the Group II Chaperonin from Thermococcus strain KS-1: Steric Hindrance by the Substituted Amino Acid, and Inter-subunit Rearrangement between Two Crystal Forms.
J.Mol.Biol., 335, 2004
|
|
4GNB
 
 | | human SMP30/GNL | | Descriptor: | CALCIUM ION, Regucalcin | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
4GN7
 
 | | mouse SMP30/GNL | | Descriptor: | CALCIUM ION, Regucalcin, SULFATE ION | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
4GN8
 
 | | mouse SMP30/GNL-1,5-AG complex | | Descriptor: | 1,5-anhydro-D-glucitol, CALCIUM ION, Regucalcin, ... | | Authors: | Aizawa, S, Senda, M, Harada, A, Maruyama, N, Ishida, T, Aigaki, T, Ishigami, A, Senda, T. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the gamma-lactone-ring formation in ascorbic acid biosynthesis by the senescence marker protein-30/gluconolactonase
Plos One, 8, 2013
|
|
2LAA
 
 | |
2LAB
 
 | |
2KTD
 
 | | Solution structure of mouse lipocalin-type prostaglandin D synthase / substrate analog (U-46619) complex | | Descriptor: | (5Z)-7-{(1R,4S,5S,6R)-6-[(1E,3S)-3-hydroxyoct-1-en-1-yl]-2-oxabicyclo[2.2.1]hept-5-yl}hept-5-enoic acid, Prostaglandin-H2 D-isomerase | | Authors: | Shimamoto, S, Maruo, H, Yoshida, T, Kato, N, Ohkubo, T. | | Deposit date: | 2010-01-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Lipocalin-type Prostaglandin D synthase / Substrate analog complex reveals Open-Closed Conformational Change required for Substrate Recognition
To be Published
|
|
2PA2
 
 | | Crystal structure of human Ribosomal protein L10 core domain | | Descriptor: | 60S ribosomal protein L10, POTASSIUM ION | | Authors: | Nishimura, M, Kaminishi, T, Takemoto, C, Kawazoe, M, Yoshida, T, Tanaka, A, Sugano, S, Shirouzu, M, Ohkubo, T, Yokoyama, S, Kobayashi, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Human Ribosomal Protein L10 Core Domain Reveals Eukaryote-Specific Motifs in Addition to the Conserved Fold
J.Mol.Biol., 377, 2008
|
|
