1BLZ
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACV-FE-NO COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Roach, P.L, Clifton, I.J, Hensgens, C.M.H, Shibata, N, Schofield, C.J, Hajdu, J, Baldwin, J.E. | | Deposit date: | 1998-07-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation.
Nature, 387, 1997
|
|
1BK0
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACV-FE COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Roach, P.L, Clifton, I.J, Hensgens, C.M.H, Shibata, N, Schofield, C.J, Hajdu, J, Baldwin, J.E. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation.
Nature, 387, 1997
|
|
1BWV
 
 | | Activated Ribulose 1,5-Bisphosphate Carboxylase/Oxygenase (RUBISCO) Complexed with the Reaction Intermediate Analogue 2-Carboxyarabinitol 1,5-Bisphosphate | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RIBULOSE BISPHOSPHATE CARBOXYLASE) | | Authors: | Sugawara, H, Yamamoto, H, Shibata, N, Inoue, T, Miyake, C, Yokota, A, Kai, Y. | | Deposit date: | 1998-09-29 | | Release date: | 1999-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of carboxylase reaction-oriented ribulose 1, 5-bisphosphate carboxylase/oxygenase from a thermophilic red alga, Galdieria partita.
J.Biol.Chem., 274, 1999
|
|
7V5P
 
 | | The dimeric structure of G80A/H81A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Xie, C, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Experimental and theoretical study on converting myoglobin into a stable domain-swapped dimer by utilizing a tight hydrogen bond network at the hinge region.
Rsc Adv, 11, 2021
|
|
1WRB
 
 | | Crystal structure of the N-terminal RecA-like domain of DjVLGB, a pranarian Vasa-like RNA helicase | | Descriptor: | DjVLGB, SULFATE ION | | Authors: | Kurimoto, K, Muto, Y, Obayashi, N, Terada, T, Shirouzu, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Kigawa, T, Okumura, H, Tanaka, A, Shibata, N, Kashikawa, M, Agata, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal RecA-like domain of a DEAD-box RNA helicase, the Dugesia japonica vasa-like gene B protein
J.Struct.Biol., 150, 2005
|
|
7DGN
 
 | | The Co-bound dimeric structure of K79H/G80A/H81A myoglobin | | Descriptor: | COBALT (II) ION, Myoglobin, OXYGEN ATOM, ... | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGJ
 
 | | The dimeric structure of K78H/G80A/H82A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGM
 
 | | The dimeric structure of K79H/G80A/H81A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGL
 
 | | The Ni-bound dimeric structure of K78H/G80A/H82A myoglobin | | Descriptor: | Myoglobin, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGO
 
 | | The Zn-bound dimeric structure of K79H/G80A/H81A myoglobin | | Descriptor: | Myoglobin, OXYGEN ATOM, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
7DGK
 
 | | The Co-bound dimeric structure of K78H/G80A/H82A myoglobin | | Descriptor: | COBALT (II) ION, Myoglobin, OXYGEN ATOM, ... | | Authors: | Nagao, S, Idomoto, A, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Rational design of metal-binding sites in domain-swapped myoglobin dimers.
J.Inorg.Biochem., 217, 2021
|
|
1WYB
 
 | | Structure of 6-aminohexanoate-dimer hydrolase | | Descriptor: | 6-aminohexanoate-dimer hydrolase | | Authors: | Negoro, S, Ohki, T, Shibata, N, Mizuno, N, Wakitani, Y, Tsurukame, J, Matsumoto, K, Kawamoto, I, Takeo, M, Higuchi, Y. | | Deposit date: | 2005-02-09 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of 6-aminohexanoate-dimer hydrolase
To be Published
|
|
1WYC
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, DN mutant | | Descriptor: | 6-aminohexanoate-dimer hydrolase | | Authors: | Negoro, S, Ohki, T, Shibata, N, Mizuno, N, Wakitani, Y, Tsurukame, J, Matsumoto, K, Kawamoto, I, Takeo, M, Higuchi, Y. | | Deposit date: | 2005-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase
J.Mol.Biol., 370, 2007
|
|
3AXG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase | | Descriptor: | Endotype 6-aminohexanoat-oligomer hydrolase, SODIUM ION | | Authors: | Negoro, S, Shibata, N, Tanaka, Y, Yasuhira, K, Shibata, H, Hashimoto, H, Lee, Y.H, Ohshima, S, Santa, R, Mochiji, K, Goto, Y, Ikegami, T, Nagai, K, Kato, D, Takeo, M, Higuchi, Y. | | Deposit date: | 2011-04-04 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
J.Biol.Chem., 287, 2012
|
|
6L1V
 
 | | Domain-swapped Alcaligenes xylosoxidans azurin dimer | | Descriptor: | Azurin-1, COPPER (II) ION | | Authors: | Cahyono, R.N, Yamanaka, M, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3D domain swapping of azurin from Alcaligenes xylosoxidans.
Metallomics, 12, 2020
|
|
5Y3C
 
 | |
5Y3B
 
 | |
6LTL
 
 | | The dimeric structure of G80A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTM
 
 | | The dimeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LS8
 
 | | The monomeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
2CVC
 
 | | Crystal structure of High-Molecular Weight Cytochrome c from Desulfovibrio vulgaris (Hildenborough) | | Descriptor: | HEME C, High-molecular-weight cytochrome c precursor | | Authors: | Suto, K, Sato, M, Shibata, N, Kitamura, M, Morimoto, Y, Takayama, Y, Ozawa, K, Akutsu, H, Higuchi, Y, Yasuoka, N. | | Deposit date: | 2005-06-02 | | Release date: | 2006-06-06 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of High-Molecular Weight Cytochrome c
To be Published
|
|
6IW3
 
 | |
6JCK
 
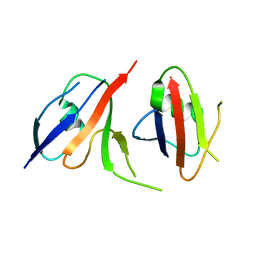 | | Complex structure of Axin-DIX and Dvl2-DIX | | Descriptor: | Axin-1, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Yamanishi, K, Shibata, N. | | Deposit date: | 2019-01-29 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | A direct heterotypic interaction between the DIX domains of Dishevelled and Axin mediates signaling to beta-catenin.
Sci.Signal., 12, 2019
|
|
6K7C
 
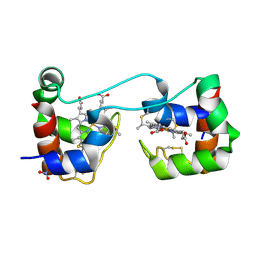 | | Dimeric Shewanella violacea cytochrome c5 | | Descriptor: | HEME C, NITRATE ION, Soluble cytochrome cA | | Authors: | Yang, H, Yamanaka, M, Nagao, S, Yasuhara, K, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Protein surface charge effect on 3D domain swapping in cells for c-type cytochromes.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
1WUH
 
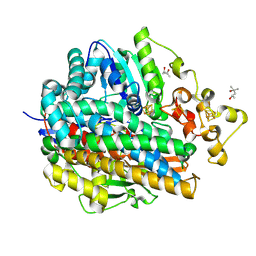 | | Three-Dimensional Structure Of The Ni-A State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Hirota, S, Nakahara, A, Komori, H, Shibata, N, Kato, T, Kano, K, Higuchi, Y. | | Deposit date: | 2004-12-07 | | Release date: | 2005-12-07 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural basis for the activation process of [NiFe] hydrogenase from D.vulgaris Miyazaki F
To be Published
|
|
