2D5M
 
 | |
1WRB
 
 | | Crystal structure of the N-terminal RecA-like domain of DjVLGB, a pranarian Vasa-like RNA helicase | | Descriptor: | DjVLGB, SULFATE ION | | Authors: | Kurimoto, K, Muto, Y, Obayashi, N, Terada, T, Shirouzu, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Kigawa, T, Okumura, H, Tanaka, A, Shibata, N, Kashikawa, M, Agata, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal RecA-like domain of a DEAD-box RNA helicase, the Dugesia japonica vasa-like gene B protein
J.Struct.Biol., 150, 2005
|
|
1WYB
 
 | | Structure of 6-aminohexanoate-dimer hydrolase | | Descriptor: | 6-aminohexanoate-dimer hydrolase | | Authors: | Negoro, S, Ohki, T, Shibata, N, Mizuno, N, Wakitani, Y, Tsurukame, J, Matsumoto, K, Kawamoto, I, Takeo, M, Higuchi, Y. | | Deposit date: | 2005-02-09 | | Release date: | 2005-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of 6-aminohexanoate-dimer hydrolase
To be Published
|
|
5Y0L
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G/H130Y mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SODIUM ION, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.385 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y0M
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D36A/D122G/H130Y/E263Q mutant | | Descriptor: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYQ
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122K mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y4N
 
 | | Crystal structure of aerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J.Inorg.Biochem., 177, 2017
|
|
6K7C
 
 | | Dimeric Shewanella violacea cytochrome c5 | | Descriptor: | HEME C, NITRATE ION, Soluble cytochrome cA | | Authors: | Yang, H, Yamanaka, M, Nagao, S, Yasuhara, K, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-06-07 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Protein surface charge effect on 3D domain swapping in cells for c-type cytochromes.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
6L1V
 
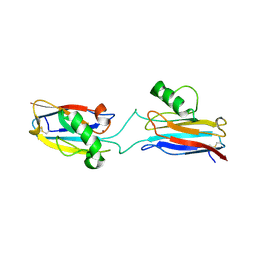 | | Domain-swapped Alcaligenes xylosoxidans azurin dimer | | Descriptor: | Azurin-1, COPPER (II) ION | | Authors: | Cahyono, R.N, Yamanaka, M, Nagao, S, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2019-09-30 | | Release date: | 2020-02-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 3D domain swapping of azurin from Alcaligenes xylosoxidans.
Metallomics, 12, 2020
|
|
5Z25
 
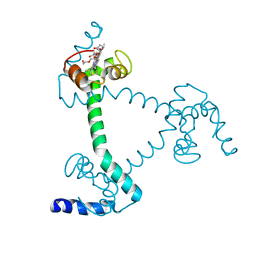 | | Trimeric Alpha-Helix-Inserted Circular Permutant of Cytochrome c555 | | Descriptor: | Cytochrome c552, HEME C, TETRAETHYLENE GLYCOL | | Authors: | Oda, A, Nagao, S, Yamanaka, M, Ueda, I, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-12-28 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Construction of a Triangle-Shaped Trimer and a Tetrahedron Using an alpha-Helix-Inserted Circular Permutant of Cytochrome c555.
Chem Asian J, 13, 2018
|
|
6LTL
 
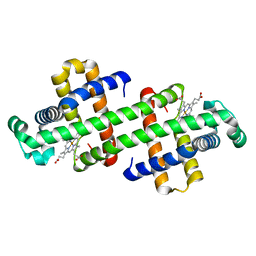 | | The dimeric structure of G80A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LTM
 
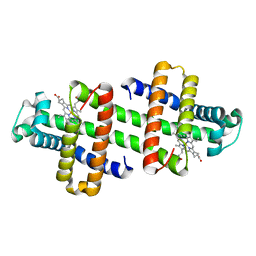 | | The dimeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6LS8
 
 | | The monomeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
5XYT
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., H130Y mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SULFATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
6JCK
 
 | | Complex structure of Axin-DIX and Dvl2-DIX | | Descriptor: | Axin-1, Segment polarity protein dishevelled homolog DVL-2 | | Authors: | Yamanishi, K, Shibata, N. | | Deposit date: | 2019-01-29 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | A direct heterotypic interaction between the DIX domains of Dishevelled and Axin mediates signaling to beta-catenin.
Sci.Signal., 12, 2019
|
|
5XYG
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72. | | Descriptor: | CHLORIDE ION, Endotype 6-aminohexanoat-oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-07 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYS
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122V mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
1WYC
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, DN mutant | | Descriptor: | 6-aminohexanoate-dimer hydrolase | | Authors: | Negoro, S, Ohki, T, Shibata, N, Mizuno, N, Wakitani, Y, Tsurukame, J, Matsumoto, K, Kawamoto, I, Takeo, M, Higuchi, Y. | | Deposit date: | 2005-02-09 | | Release date: | 2006-02-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Nylon-oligomer degrading enzyme/substrate complex: catalytic mechanism of 6-aminohexanoate-dimer hydrolase
J.Mol.Biol., 370, 2007
|
|
3A66
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N/D370Y mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOATE-DIMER HYDROLASE, 6-AMINOHEXANOIC ACID, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
3A65
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/H266N mutant with substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2009-08-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enzymatic Synthesis of Nylon-6 Units in Organic Sol Contained Low-Water: Structural Requirement of 6-Aminohexanoate-Dimer Hydrolase for Efficient Amid Synthesis
To be Published
|
|
2ZM8
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/D370Y Mutant Complexed with 6-Aminohexanoate-dimer | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZMA
 
 | | Crystal Structure of 6-Aminohexanoate-dimer Hydrolase S112A/G181D/H266N/D370Y Mutant with Substrate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Takeo, M, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
2ZM7
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/G181D Mutant Complexed with 6-Aminohexanoate-dimer | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular design of a nylon-6 byproduct-degrading enzyme from a carboxylesterase with a beta-lactamase fold
Febs J., 276, 2009
|
|
3VQT
 
 | | Crystal structure analysis of the translation factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor 3 | | Authors: | Kihira, K, Shomura, Y, Shibata, N, Kitamura, M, Higuchi, Y. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of the translation factor RF3 (release factor 3)
Febs Lett., 586, 2012
|
|
3VWN
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187G/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
