3W8M
 
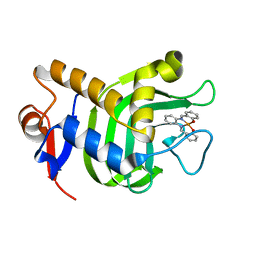 | | Crystal Structure of HasAp with Iron Salophen | | Descriptor: | 2,2'-[1,2-PHENYLENEBIS(NITRILOMETHYLIDYNE)]BIS[PHENOLATO]](2-)-N,N',O,O']-IRON, Heme acquisition protein HasAp | | Authors: | Shirataki, C, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-03-19 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3W8O
 
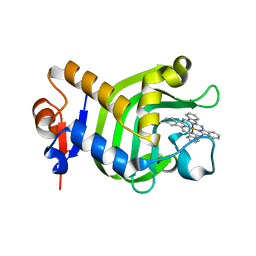 | | Crystal Structure of HasAp with Iron Phthalocyanine | | Descriptor: | Heme acquisition protein HasAp, [29H,31H-phthalocyaninato(2-)-kappa~4~N~29~,N~30~,N~31~,N~32~]iron | | Authors: | Shirataki, C, Shoji, O, Terada, M, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-03-20 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2III
 
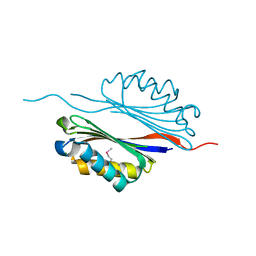 | | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, S-adenosylmethionine decarboxylase proenzyme | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the adenosylmethionine decarboxylase (aq_254) from aquifex aeolicus vf5
To be Published
|
|
6JLG
 
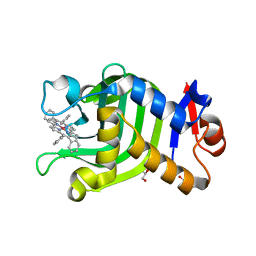 | | Crystal Structure of HasAp with Co-9,10,19,20-Tetraphenylporphycene | | Descriptor: | GLYCEROL, Heme acquisition protein HasA, PHOSPHATE ION, ... | | Authors: | Sakakibara, E, Shisaka, Y, Onoda, H, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Highly malleable haem-binding site of the haemoprotein HasA permits stable accommodation of bulky tetraphenylporphycenes.
Rsc Adv, 9, 2019
|
|
3WAH
 
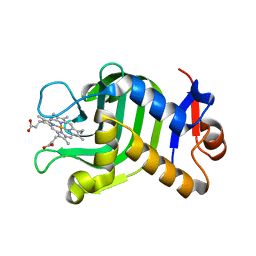 | | Crystal Structure of HasAp with Iron MesoporphyrinIX | | Descriptor: | Heme acquisition protein HasAp, Mesoheme | | Authors: | Shirataki, C, Shoji, O, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2013-05-02 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Inhibition of Heme Uptake in Pseudomonas aeruginosa by its Hemophore (HasAp ) Bound to Synthetic Metal Complexes
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2IIH
 
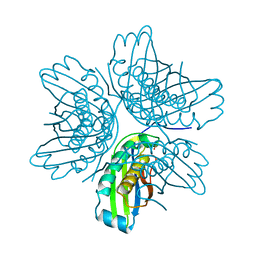 | | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form) | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Chen, L, Liu, Z.-J, Wang, B.-C, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-28 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from thermus theromophilus HB8 (H32 form)
To be Published
|
|
2IS8
 
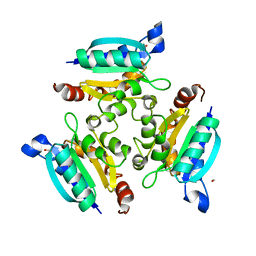 | | Crystal structure of the Molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8 | | Descriptor: | FORMIC ACID, Molybdopterin biosynthesis enzyme, MoaB | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of the molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8
To be Published
|
|
2IRP
 
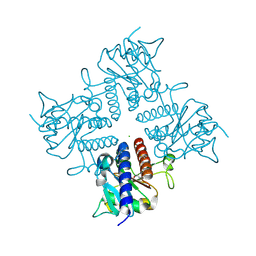 | | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Putative aldolase class 2 protein aq_1979 | | Authors: | Jeyakanthan, J, Gayathri, D, Yogavel, M, Velmurugan, D, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-16 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the l-fuculose-1-phosphate aldolase (aq_1979) from aquifex aeolicus VF5
To be Published
|
|
8JS5
 
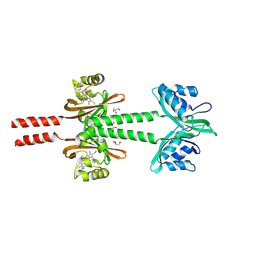 | | Dimeric PAS domains of oxygen sensor FixL with ferric unliganded heme | | Descriptor: | GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, Sensor protein FixL | | Authors: | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
8JS7
 
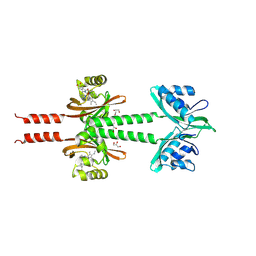 | | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme | | Descriptor: | GLYCEROL, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
8JS6
 
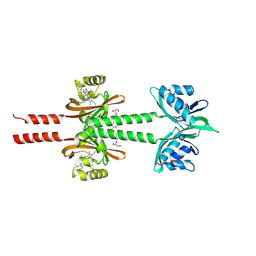 | | Dimeric PAS domains of oxygen sensor FixL in complex with cyanide-bound ferric heme | | Descriptor: | CYANIDE ION, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kamaya, M, Koteishi, H, Sawai, H, Sugimoto, H, Shiro, Y. | | Deposit date: | 2023-06-19 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dimeric PAS domains of oxygen sensor FixL in complex with imidazole-bound heme.
To be published
|
|
1JF6
 
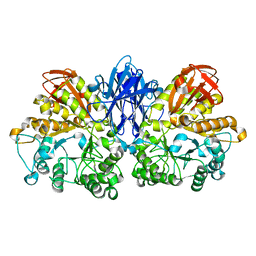 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
2IDE
 
 | | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8
To be Published
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
1JF5
 
 | | CRYSTAL STRUCTURE OF THERMOACTINOMYCES VULGARIS R-47 ALPHA-AMYLASE 2 MUTANT F286A | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
2KFJ
 
 | | Solution structure of the loop deletion mutant of PB1 domain of Cdc24p | | Descriptor: | Cell division control protein 24 | | Authors: | Ogura, K, Tandai, T, Yoshinaga, S, Kobashigawa, Y, Kumeta, H, Inagaki, F. | | Deposit date: | 2009-02-22 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the heterodimer of Bem1 and Cdc24 PB1 domains from Saccharomyces cerevisiae
J.Biochem., 146, 2009
|
|
1UG2
 
 | | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain | | Descriptor: | 2610100B20Rik gene product | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain
To be Published
|
|
2IEX
 
 | | Crystal structure of dihydroxynapthoic acid synthetase (GK2873) from Geobacillus kaustophilus HTA426 | | Descriptor: | Dihydroxynapthoic acid synthetase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, BaBa, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-19 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of dihydroxynapthoic acid synthetase (GK2873) from Geobacillus kaustophilus HTA426
To be Published
|
|
1CL6
 
 | |
4GRV
 
 | | The crystal structure of the neurotensin receptor NTS1 in complex with neurotensin (8-13) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Neurotensin 8-13, Neurotensin receptor type 1, ... | | Authors: | Noinaj, N, White, J.F, Shibata, Y, Love, J, Kloss, B, Xu, F, Gvozdenovic-Jeremic, J, Shah, P, Shiloach, J, Tate, C.G, Grisshammer, R. | | Deposit date: | 2012-08-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure of the agonist-bound neurotensin receptor.
Nature, 490, 2012
|
|
1CMN
 
 | |
2HOQ
 
 | |
4BGN
 
 | | cryo-EM structure of the NavCt voltage-gated sodium channel | | Descriptor: | VOLTAGE-GATED SODIUM CHANNEL | | Authors: | Tsai, C.J, Tani, K, Irie, K, Hiroaki, Y, Shimomura, T, Mcmillan, D.G, Cook, G.M, Schertler, G, Fujiyoshi, Y, Li, X.D. | | Deposit date: | 2013-03-28 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | ELECTRON CRYSTALLOGRAPHY (9 Å) | | Cite: | Two Alternative Conformations of a Voltage-Gated Sodium Channel.
J.Mol.Biol., 425, 2013
|
|
7DHL
 
 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | Descriptor: | 5-[2-(3,5-dimethoxyphenyl)ethyl]-N-[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]pyrimidin-2-amine, Fibroblast growth factor receptor 3 | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Synthesis and structure-activity relationships of pyrimidine derivatives as potent and orally active FGFR3 inhibitors with both increased systemic exposure and enhanced in vitro potency.
Bioorg.Med.Chem., 33, 2021
|
|
1JIB
 
 | | Complex of Alpha-amylase II (TVA II) from Thermoactinomyces vulgaris R-47 with Maltotetraose Based on a Crystal Soaked with Maltohexaose. | | Descriptor: | NEOPULLULANASE, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yokota, T, Tonozuka, T, Shimura, Y, Ichikawa, K, Kamitori, S, Sakano, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2001-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Thermoactinomyces vulgaris R-47 alpha-amylase II complexed with substrate analogues.
Biosci.Biotechnol.Biochem., 65, 2001
|
|
