7BOM
 
 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr immobilized with gold ions. | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
7BON
 
 | | Crystal structure of recombinant horse spleen apo-R52C/E56C/R59C/E63C-Fr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Maity, B, Ito, N, Abe, S, Lu, D, Ueno, T. | | Deposit date: | 2020-03-19 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Design of Multinuclear Gold Binding Site at the Two-fold Symmetric Interface of the Ferritin Cage
Chem Lett., 49, 2021
|
|
7YBC
 
 | |
7YB8
 
 | |
7YBA
 
 | |
7YB9
 
 | |
7YBB
 
 | |
7R8J
 
 | |
7R8F
 
 | |
7R8K
 
 | |
7R8G
 
 | |
7R8H
 
 | |
7R8I
 
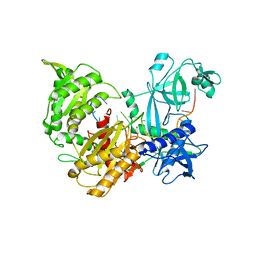 | |
4ZID
 
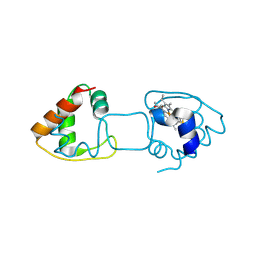 | | Dimeric Hydrogenobacter thermophilus cytochrome c552 obtained from Escherichia coli | | Descriptor: | Cytochrome c-552, HEME C | | Authors: | Hayashi, Y, Yamanaka, M, Nagao, S, Komori, H, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-04-28 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Domain swapping oligomerization of thermostable c-type cytochrome in E. coli cells
Sci Rep, 6, 2016
|
|
7SZK
 
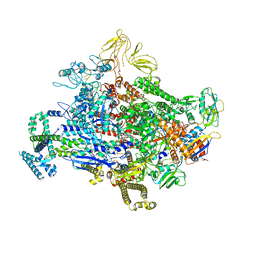 | | Cryo-EM structure of 27a bound to E. coli RNAP and rrnBP1 promoter complex | | Descriptor: | (2S,7R,7aR,13aP,16Z,18E,20S,21S,22R,23R,24R,25S,26R,27S,28E)-5,21,23-trihydroxy-27-methoxy-2,4,16,20,22,24,26-heptamethyl-10-[4-(2-methylpropyl)piperazin-1-yl]-12-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1,6,15-trioxo-1,2,7,7a-tetrahydro-6H-2,7-(epoxypentadeca[1,11,13]trienoimino)[1]benzofuro[4,5-a]phenoxazin-25-yl acetate, DNA (5'-D(P*CP*TP*CP*GP*TP*AP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shin, Y, Murakami, K.S. | | Deposit date: | 2021-11-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Optimization of Benzoxazinorifamycins to Improve Mycobacterium tuberculosis RNA Polymerase Inhibition and Treatment of Tuberculosis.
Acs Infect Dis., 8, 2022
|
|
7SZJ
 
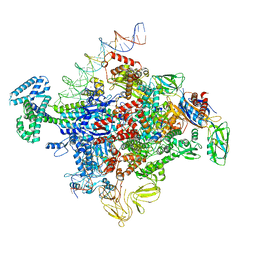 | |
1QPK
 
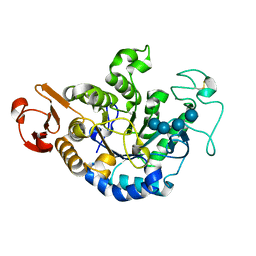 | | MUTANT (D193G) MALTOTETRAOSE-FORMING EXO-AMYLASE IN COMPLEX WITH MALTOTETRAOSE | | Descriptor: | CALCIUM ION, PROTEIN (MALTOTETRAOSE-FORMING AMYLASE), alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1999-05-26 | | Release date: | 1999-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of catalytic residues in alpha-amylases as evidenced by the structures of the product-complexed mutants of a maltotetraose-forming amylase.
Protein Eng., 12, 1999
|
|
3KDO
 
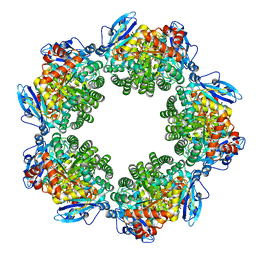 | | Crystal structure of Type III Rubisco SP6 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
3KDN
 
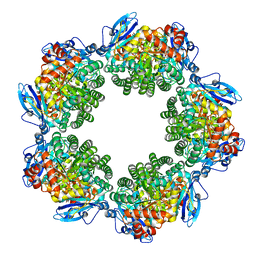 | | Crystal structure of Type III Rubisco SP4 mutant complexed with 2-CABP | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase | | Authors: | Nishitani, Y, Fujihashi, M, Doi, T, Yoshida, S, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-based catalytic optimization of a type III Rubisco from a hyperthermophile
J.Biol.Chem., 285, 2010
|
|
5HEE
 
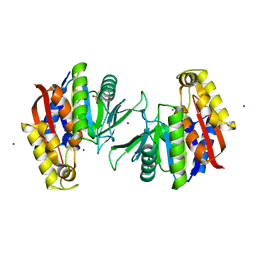 | | Crystal structure of the TK2203 protein | | Descriptor: | GLYCEROL, Putative uncharacterized protein, TK2203 protein, ... | | Authors: | Nishitani, Y, Miki, K. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
3O26
 
 | |
2RSE
 
 | | NMR structure of FKBP12-mTOR FRB domain-rapamycin complex structure determined based on PCS | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Serine/threonine-protein kinase mTOR, TERBIUM(III) ION | | Authors: | Kobashigawa, Y, Ushio, M, Saio, T, Inagaki, F. | | Deposit date: | 2012-01-25 | | Release date: | 2012-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Convenient method for resolving degeneracies due to symmetry of the magnetic susceptibility tensor and its application to pseudo contact shift-based protein-protein complex structure determination.
J.Biomol.Nmr, 53, 2012
|
|
1JDC
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 1) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
1JDD
 
 | | MUTANT (E219Q) MALTOTETRAOSE-FORMING EXO-AMYLASE COCRYSTALLIZED WITH MALTOTETRAOSE (CRYSTAL TYPE 2) | | Descriptor: | 1,4-ALPHA MALTOTETRAHYDROLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Yoshioka, Y, Hasegawa, K, Matsuura, Y, Katsube, Y, Kubota, M. | | Deposit date: | 1997-06-16 | | Release date: | 1997-10-15 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a mutant maltotetraose-forming exo-amylase cocrystallized with maltopentaose.
J.Mol.Biol., 271, 1997
|
|
