8K72
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((3-(phenylsulfonamido)propanoyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | 分子名称: | 2-[(2~{Z})-3-oxidanyl-2-[3-(phenylsulfonylamino)propanoylimino]-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | 著者 | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | 登録日 | 2023-07-26 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
8K73
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((1-(phenylsulfonyl)pyrrolidine-3-carbonyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | 分子名称: | 2-[(2~{Z})-3-oxidanyl-2-[(3~{R})-1-(phenylsulfonyl)pyrrolidin-3-yl]carbonylimino-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | 著者 | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | 登録日 | 2023-07-26 | | 公開日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
8KI0
 
 | | Crystal structure of the hemophore HasA from Pseudomonas protegens Pf-5 capturing Fe-tetraphenylporphyrin | | 分子名称: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Heme acquisition protein HasAp, ... | | 著者 | Shisaka, Y, Inaba, H, Sugimoto, H, Shoji, O. | | 登録日 | 2023-08-22 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom.
Rsc Adv, 14, 2024
|
|
8KI1
 
 | | Crystal structure of the holo form of the hemophore HasA from Pseudomonas protegens Pf-5 | | 分子名称: | GLYCEROL, Heme acquisition protein HasAp, PHOSPHATE ION, ... | | 著者 | Shisaka, Y, Inaba, H, Sugimoto, H, Shoji, O. | | 登録日 | 2023-08-22 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom.
Rsc Adv, 14, 2024
|
|
8JX2
 
 | | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer | | 分子名称: | alpha hemolysin fused with spy-catcher, alpha hemolysin fused with spy-tag | | 著者 | Ishii, Y, Naito, K, Yokoyama, T, Tanaka, Y, Matsuura, T. | | 登録日 | 2023-06-30 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer
To Be Published
|
|
4GA4
 
 | | Crystal structure of AMP phosphorylase N-terminal deletion mutant | | 分子名称: | PHOSPHATE ION, Putative thymidine phosphorylase | | 著者 | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2012-07-25 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.51 Å) | | 主引用文献 | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
8JX3
 
 | | alpha-Hemolysin(G122S/K147R/K237C)-SpyTag/SpyCatcher head to head 14-mer | | 分子名称: | alpha hemolysin fused with spy-catcher, alpha hemolysin fused with spy-tag | | 著者 | Ishii, Y, Naito, K, Yokoyama, T, Tanaka, Y, Matsuura, T. | | 登録日 | 2023-06-30 | | 公開日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.2 Å) | | 主引用文献 | alpha-Hemolysin(G122S/K147R)-SpyTag/SpyCatcher head to head 14-mer
To Be Published
|
|
4GA5
 
 | | Crystal structure of AMP phosphorylase C-terminal deletion mutant in the apo-form | | 分子名称: | Putative thymidine phosphorylase | | 著者 | Nishitani, Y, Aono, R, Nakamura, A, Sato, T, Atomi, H, Imanaka, T, Miki, K. | | 登録日 | 2012-07-25 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.25 Å) | | 主引用文献 | Structure analysis of archaeal AMP phosphorylase reveals two unique modes of dimerization
J.Mol.Biol., 425, 2013
|
|
8ITH
 
 | |
8ITG
 
 | |
4GPG
 
 | | X/N joint refinement of Achromobacter Lyticus Protease I free form at pD8.0 | | 分子名称: | Protease 1 | | 著者 | Ohnishi, Y, Yamada, T, Kurihara, K, Tanaka, I, Sakiyama, F, Masaki, T, Niimura, N. | | 登録日 | 2012-08-21 | | 公開日 | 2013-09-11 | | 最終更新日 | 2023-11-08 | | 実験手法 | NEUTRON DIFFRACTION (1.895 Å), X-RAY DIFFRACTION | | 主引用文献 | Neutron and X-ray crystallographic analysis of Achromobacter protease I at pD 8.0: protonation states and hydration structure in the free-form.
Biochim.Biophys.Acta, 1834, 2013
|
|
7F8D
 
 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) G218Y mutant | | 分子名称: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | 登録日 | 2021-07-02 | | 公開日 | 2022-02-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Increasing loop flexibility affords low-temperature adaptation of a moderate thermophilic malate dehydrogenase from Geobacillus stearothermophilus.
Protein Eng.Des.Sel., 34, 2021
|
|
1I9Y
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN | | 分子名称: | PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | 著者 | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | 登録日 | 2001-03-21 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I9Z
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 5-PHOSPHATASE DOMAIN (IPP5C) OF SPSYNAPTOJANIN IN COMPLEX WITH INOSITOL (1,4)-BISPHOSPHATE AND CALCIUM ION | | 分子名称: | CALCIUM ION, D-MYO-INOSITOL-1,4-BISPHOSPHATE, PHOSPHATIDYLINOSITOL PHOSPHATE PHOSPHATASE | | 著者 | Tsujishita, Y, Guo, S, Stolz, L, York, J.D, Hurley, J.H. | | 登録日 | 2001-03-21 | | 公開日 | 2001-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase.
Cell(Cambridge,Mass.), 105, 2001
|
|
1I56
 
 | |
1J35
 
 | | Crystal Structure of Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein | | 分子名称: | CALCIUM ION, Coagulation factor IX, Coagulation factor IX-binding protein B chain, ... | | 著者 | Shikamoto, Y, Morita, T, Fujimoto, Z, Mizuno, H. | | 登録日 | 2003-01-20 | | 公開日 | 2003-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Mg2+- and Ca2+-bound Gla Domain of Factor IX Complexed with Binding Protein
J.Biol.Chem., 278, 2003
|
|
8H8N
 
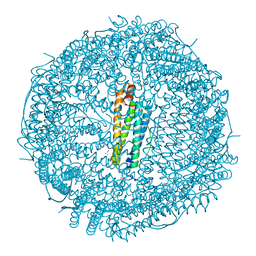 | | Crystal structure of apo-R52Y/E56Y/R59Y/E63Y-rHLFr | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | 登録日 | 2022-10-23 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8L
 
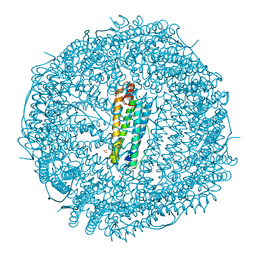 | | Crystal structure of apo-R52F/E56F/R59F/E63F-rHLFr | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | 登録日 | 2022-10-23 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8M
 
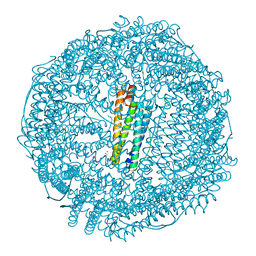 | | Crystal structure of apo-E53F/E57F/E60F/E64F-rHLFr | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | 登録日 | 2022-10-23 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8O
 
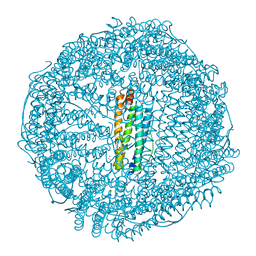 | | Crystal structure of apo-R52W/E56W/R59W/E63W-rHLFr | | 分子名称: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | 登録日 | 2022-10-23 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
1J34
 
 | | Crystal Structure of Mg(II)-and Ca(II)-bound Gla Domain of Factor IX Complexed with Binding Protein | | 分子名称: | CALCIUM ION, Coagulation factor IX, MAGNESIUM ION, ... | | 著者 | Shikamoto, Y, Morita, T, Fujimoto, Z, Mizuno, H. | | 登録日 | 2003-01-20 | | 公開日 | 2003-07-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal Structure of Mg2+- and Ca2+-bound Gla Domain of Factor IX Complexed with Binding Protein
J.Biol.Chem., 278, 2003
|
|
5HEE
 
 | | Crystal structure of the TK2203 protein | | 分子名称: | GLYCEROL, Putative uncharacterized protein, TK2203 protein, ... | | 著者 | Nishitani, Y, Miki, K. | | 登録日 | 2016-01-06 | | 公開日 | 2016-06-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Crystal structure of the TK2203 protein from Thermococcus kodakarensis, a putative extradiol dioxygenase
Acta Crystallogr.,Sect.F, 72, 2016
|
|
6KV0
 
 | | Ferredoxin I from C. reinhardtii, high X-ray dose | | 分子名称: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Onishi, Y, Kurisu, G, Tanaka, H. | | 登録日 | 2019-09-03 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6KUM
 
 | | Ferredoxin I from C. reinhardtii, low X-ray dose | | 分子名称: | BENZAMIDINE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | 著者 | Onishi, Y, Kurisu, G, Tanaka, H. | | 登録日 | 2019-09-02 | | 公開日 | 2020-05-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
6LK1
 
 | | Ultrahigh resolution X-ray structure of Ferredoxin I from C. reinhardtii | | 分子名称: | BENZAMIDINE, FE2/S2 (INORGANIC) CLUSTER, Ferredoxin, ... | | 著者 | Onishi, Y, Kurisu, G, Tanaka, H. | | 登録日 | 2019-12-17 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | X-ray dose-dependent structural changes of the [2Fe-2S] ferredoxin from Chlamydomonas reinhardtii.
J.Biochem., 167, 2020
|
|
