5X5B
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 2 | | 分子名称: | Spike glycoprotein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
2RSN
 
 | |
1ETL
 
 | |
1ETM
 
 | |
5X59
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, three-fold symmetry | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
2RVM
 
 | |
3VHI
 
 | | Crystal structure of monoZ-biotin-avidin complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-{(3aS,4S,6aR)-1-[(benzyloxy)carbonyl]-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl}pentanoic acid, Avidin, ... | | 著者 | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | 登録日 | 2011-08-25 | | 公開日 | 2011-12-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
2RPV
 
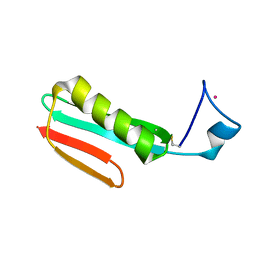 | | Solution Structure of GB1 with LBT probe | | 分子名称: | Immunoglobulin G-binding protein G, LANTHANUM (III) ION | | 著者 | Saio, T, Ogura, K, Yokochi, M, Kobashigawa, Y, Inagaki, F. | | 登録日 | 2008-10-28 | | 公開日 | 2009-09-15 | | 最終更新日 | 2021-11-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two-point anchoring of a lanthanide-binding peptide to a target protein enhances the paramagnetic anisotropic effect
J.Biomol.Nmr, 44, 2009
|
|
2RVB
 
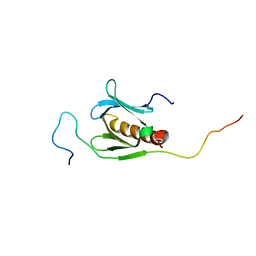 | |
2RNF
 
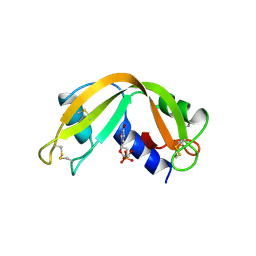 | | X-RAY CRYSTAL STRUCTURE OF HUMAN RIBONUCLEASE 4 IN COMPLEX WITH D(UP) | | 分子名称: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, RIBONUCLEASE 4 | | 著者 | Terzyan, S.S, Peracaula, R, De Llorens, R, Tsushima, Y, Yamada, H, Seno, M, Gomis-Ruth, F.X, Coll, M. | | 登録日 | 1998-11-03 | | 公開日 | 1999-11-10 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The three-dimensional structure of human RNase 4, unliganded and complexed with d(Up), reveals the basis for its uridine selectivity.
J.Mol.Biol., 285, 1999
|
|
5X5F
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 2 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X4S
 
 | | Structure of the N-terminal domain (NTD)of SARS-CoV spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | 著者 | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | 登録日 | 2017-02-14 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X58
 
 | | Prefusion structure of SARS-CoV spike glycoprotein, conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
5X5C
 
 | | Prefusion structure of MERS-CoV spike glycoprotein, conformation 1 | | 分子名称: | S protein | | 著者 | Yuan, Y, Cao, D, Zhang, Y, Ma, J, Qi, J, Wang, Q, Lu, G, Wu, Y, Yan, J, Shi, Y, Zhang, X, Gao, G.F. | | 登録日 | 2017-02-15 | | 公開日 | 2017-05-03 | | 最終更新日 | 2017-05-24 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
3VS9
 
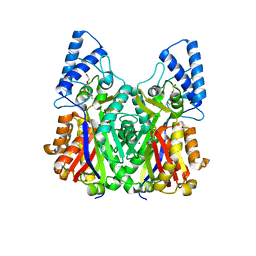 | | Crystal structure of type III PKS ArsC mutant | | 分子名称: | SODIUM ION, TETRAETHYLENE GLYCOL, Type III polyketide synthase | | 著者 | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | 登録日 | 2012-04-23 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
1EWK
 
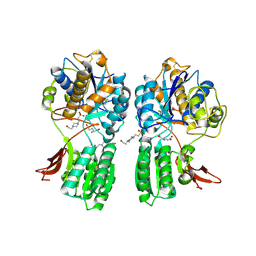 | | CRYSTAL STRUCTURE OF METABOTROPIC GLUTAMATE RECEPTOR SUBTYPE 1 COMPLEXED WITH GLUTAMATE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTAMIC ACID, ... | | 著者 | Kunishima, N, Shimada, Y, Jingami, H, Morikawa, K. | | 登録日 | 2000-04-26 | | 公開日 | 2000-12-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor.
Nature, 407, 2000
|
|
3VS8
 
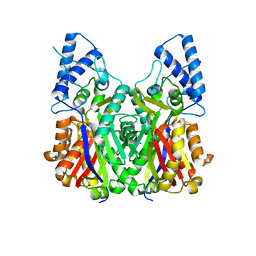 | | Crystal structure of type III PKS ArsC | | 分子名称: | SODIUM ION, Type III polyketide synthase | | 著者 | Satou, R, Miyanaga, A, Ozawa, H, Funa, N, Miyazono, K, Tanokura, M, Ohnishi, Y, Horinouchi, S. | | 登録日 | 2012-04-23 | | 公開日 | 2013-04-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Structural basis for cyclization specificity of two Azotobacter type III polyketide synthases: a single amino acid substitution reverses their cyclization specificity
J.Biol.Chem., 288, 2013
|
|
3VOU
 
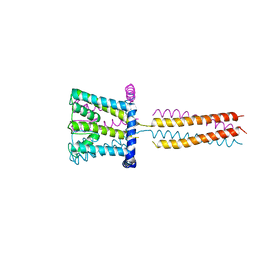 | | The crystal structure of NaK-NavSulP chimera channel | | 分子名称: | COBALT (II) ION, Ion transport 2 domain protein, Voltage-gated sodium channel, ... | | 著者 | Irie, K, Shimomura, T, Fujiyoshi, Y. | | 登録日 | 2012-02-10 | | 公開日 | 2012-05-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | The C-terminal helical bundle of the tetrameric prokaryotic sodium channel accelerates the inactivation rate
Nat Commun, 3, 2012
|
|
5XLO
 
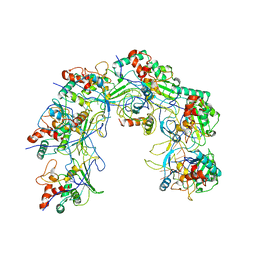 | |
5XGR
 
 | | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1 | | 著者 | Xue, H, Qi, J, Song, H, Qihui, W, Shi, Y, Gao, G.F. | | 登録日 | 2017-04-16 | | 公開日 | 2017-05-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the S1 subunit C-terminal domain from bat-derived coronavirus HKU5 spike protein
Virology, 507, 2017
|
|
5XLP
 
 | |
1SE0
 
 | | Crystal structure of DIAP1 BIR1 bound to a Grim peptide | | 分子名称: | Apoptosis 1 inhibitor, Cell death protein Grim, ZINC ION | | 著者 | Yan, N, Wu, J.W, Shi, Y. | | 登録日 | 2004-02-15 | | 公開日 | 2004-04-27 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TY4
 
 | | Crystal structure of a CED-9/EGL-1 complex | | 分子名称: | Apoptosis regulator ced-9, EGg Laying defective EGL-1, programmed cell death activator | | 著者 | Yan, N, Gu, L, Kokel, D, Xue, D, Shi, Y. | | 登録日 | 2004-07-07 | | 公開日 | 2004-09-28 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural, Biochemical, and Functional Analyses of CED-9 Recognition by the Proapoptotic Proteins EGL-1 and CED-4
Mol.Cell, 15, 2004
|
|
5YYZ
 
 | | Crystal structure of the MEK1 FHA domain in complex with the HOP1 pThr318 peptide. | | 分子名称: | Meiosis-specific protein HOP1, Meiosis-specific serine/threonine-protein kinase MEK1 | | 著者 | Xie, C, Li, F, Jiang, Y, Wu, J, Shi, Y. | | 登録日 | 2017-12-11 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.798 Å) | | 主引用文献 | Structural insights into the recognition of phosphorylated Hop1 by Mek1
Acta Crystallogr D Struct Biol, 74, 2018
|
|
