3WU3
 
 | | Reduced-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3WU4
 
 | | Oxidized-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
1UJD
 
 | | Solution Structure of RSGI RUH-003, a PDZ domain of hypothetical KIAA0559 protein from human cDNA | | Descriptor: | KIAA0559 protein | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-31 | | Release date: | 2004-01-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-003, a PDZ domain of hypothetical KIAA0559 protein from human cDNA
To be published
|
|
2CP9
 
 | | Solution structure of RSGI RUH-042, a UBA domain from human mitochondrial elongation factor Ts | | Descriptor: | Elongation factor Ts, mitochondrial | | Authors: | Ohashi, W, Hirota, H, Izumi, K, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-042, a UBA domain from human mitochondrial elongation factor Ts
To be Published
|
|
1WIX
 
 | | The solution structure of RSGI RUH-026, conserved domain of HOOK1 protein from mouse | | Descriptor: | Hook homolog 1 | | Authors: | Ohashi, W, Yamazaki, T, Hirota, H, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of RSGI RUH-026, conserved domain of HOOK1 protein from mouse
To be Published
|
|
2CP8
 
 | | Solution structure of the RSGI RUH-046, a UBA domain from human Next to BRCA1 gene 1 protein (KIAA0049 protein) R923H variant | | Descriptor: | Next to BRCA1 gene 1 protein | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-19 | | Release date: | 2005-11-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RSGI RUH-046, a UBA domain from human Next to BRCA1 gene 1 protein (KIAA0049 protein) R923H variant
To be Published
|
|
6OBU
 
 | | PP1 Y134K in complex with Microcystin LR | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBN
 
 | | The crystal structure of coexpressed SDS22:PP1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OBR
 
 | | PP1 Y134A in complex with Microcystin LR | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Microcystin LR, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O8A
 
 | | Thaumatin native-SAD structure determined at 5 keV from microcrystals | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Guo, G, Zhu, P, Fuchs, M.R, Shi, W, Andi, B, Gao, Y, Hendrickson, W.A, McSweeney, S, Liu, Q. | | Deposit date: | 2019-03-09 | | Release date: | 2019-05-08 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synchrotron microcrystal native-SAD phasing at a low energy.
Iucrj, 6, 2019
|
|
8D56
 
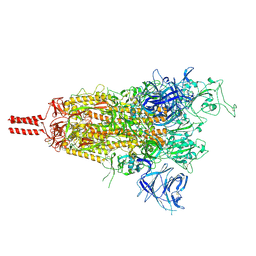 | | One RBD-up state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D55
 
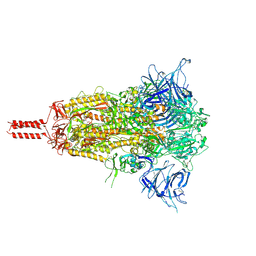 | | Closed state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D5A
 
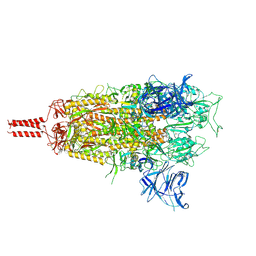 | | Middle state of SARS-CoV-2 BA.2 variant spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Tang, W.C, Gao, H.L, Shi, W, Peng, H.Q, Volloch, S.R, Xiao, T.S, Chen, B. | | Deposit date: | 2022-06-04 | | Release date: | 2023-06-07 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and functional characteristics of the SARS-CoV-2 Omicron subvariant BA.2 spike protein.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6VCG
 
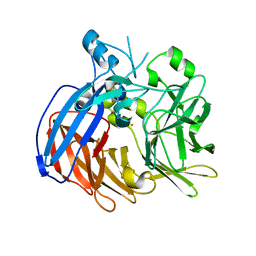 | | Crystal structure of Nitrosotalea devanaterra carotenoid cleavage dioxygenase, cobalt form | | Descriptor: | CHLORIDE ION, COBALT (II) ION, SODIUM ION, ... | | Authors: | Daruwalla, A, Shi, W, Kiser, P.D. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for carotenoid cleavage by an archaeal carotenoid dioxygenase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6OBS
 
 | | PP1 Y134K | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6XL9
 
 | | Cryo-EM structure of EcmrR-RNAP-promoter initial transcribing complex with 3-nt RNA transcript (EcmrR-RPitc-3nt) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XLA
 
 | | Cryo-EM structure of EcmrR-DNA complex in EcmrR-RPitc-3nt | | Descriptor: | MerR family transcriptional regulator EcmrR, TETRAPHENYLANTIMONIUM ION, synthetic non-template strand DNA (54-MER), ... | | Authors: | Yang, Y, Liu, C, Shi, W, Liu, B. | | Deposit date: | 2020-06-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of transcription activated by a multidrug-sensing MerR family regulator.
Nat Commun, 12, 2021
|
|
6XH7
 
 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XH8
 
 | | CueR-transcription activation complex with RNA transcript | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6PMJ
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 2 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
6PMI
 
 | | Sigm28-transcription initiation complex with specific promoter at the state 1 | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Liu, B, Shi, W. | | Deposit date: | 2019-07-02 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of bacterial sigma28-mediated transcription reveals roles of the RNA polymerase zinc-binding domain.
Embo J., 39, 2020
|
|
1B8O
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fedorov, A.A, Kicska, G.A, Fedorov, E.V, Shi, W, Tyler, P.C, Furneaux, R.H, Schramm, V.L, Almo, S.C. | | Deposit date: | 1999-02-02 | | Release date: | 1999-02-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Transition state structure of purine nucleoside phosphorylase and principles of atomic motion in enzymatic catalysis.
Biochemistry, 40, 2001
|
|
1A3W
 
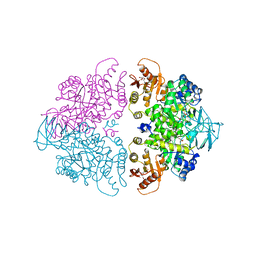 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH FBP, PG, MN2+ AND K+ | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
1A3X
 
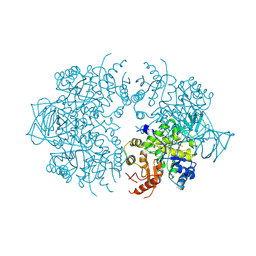 | | PYRUVATE KINASE FROM SACCHAROMYCES CEREVISIAE COMPLEXED WITH PG, MN2+ AND K+ | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, POTASSIUM ION, ... | | Authors: | Jurica, M.S, Mesecar, A, Heath, P.J, Shi, W, Nowak, T, Stoddard, B.L. | | Deposit date: | 1998-01-26 | | Release date: | 1998-05-27 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate.
Structure, 6, 1998
|
|
6XQB
 
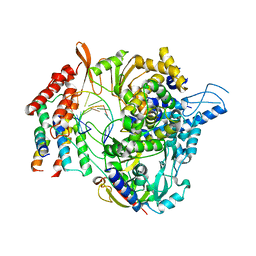 | | SARS-CoV-2 RdRp/RNA complex | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-07-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of SARS-CoV-2 RdRp/RNA complex at 3.4 Angstroms resolution
To Be Published
|
|
