2FV0
 
 | | UGL_D88N/dGlcA-Glc-Rha-Glc | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-beta-D-glucopyranose-(1-4)-alpha-L-rhamnopyranose-(1-3)-beta-D-glucopyranose, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
2FV1
 
 | | UGL_D88N/dGlcA-GlcNAc | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Unsaturated glucuronyl hydrolase | | Authors: | Itoh, T, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2006-01-28 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Substrate recognition by unsaturated glucuronyl hydrolase from Bacillus sp. GL1
Biochem.Biophys.Res.Commun., 344, 2006
|
|
7WGU
 
 | | Crystal structure of metal-binding protein EfeO from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Iron uptake system protein EfeO, ... | | Authors: | Nakatsuji, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-12-29 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of EfeB and EfeO in a bacterial siderophore-independent iron transport system
Biochem.Biophys.Res.Commun., 594, 2022
|
|
2K3R
 
 | | Pfu Rpp21 structure and assignments | | Descriptor: | Ribonuclease P protein component 4, ZINC ION | | Authors: | Amero, C.D, Foster, M.P, Boomershine, W.P, Xu, Y. | | Deposit date: | 2008-05-15 | | Release date: | 2008-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Pyrococcus furiosus RPP21, a component of the archaeal RNase P holoenzyme, and interactions with its RPP29 protein partner.
Biochemistry, 47, 2008
|
|
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1HV6
 
 | | CRYSTAL STRUCTURE OF ALGINATE LYASE A1-III COMPLEXED WITH TRISACCHARIDE PRODUCT. | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-D-glucopyranuronic acid, ALGINATE LYASE, SULFATE ION | | Authors: | Yoon, H.-J, Hashimoto, W, Miyake, O, Murata, K, Mikami, B. | | Deposit date: | 2001-01-08 | | Release date: | 2001-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of alginate lyase A1-III complexed with trisaccharide product at 2.0 A resolution.
J.Mol.Biol., 307, 2001
|
|
6JBN
 
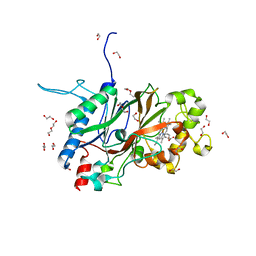 | | Crystal structure of Sphingomonas sp. A1 peroxidase EfeB responsible for import of iron | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, OXYGEN MOLECULE, ... | | Authors: | Okumura, K, Takase, R, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rare metal binding by a cell-surface component of bacterial EfeUOB iron importer
To Be Published
|
|
1FP3
 
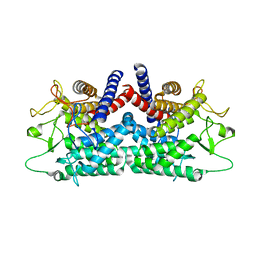 | | CRYSTAL STRUCTURE OF N-ACYL-D-GLUCOSAMINE 2-EPIMERASE FROM PORCINE KIDNEY | | Descriptor: | N-ACYL-D-GLUCOSAMINE 2-EPIMERASE | | Authors: | Itoh, T, Mikami, B, Maru, I, Ohta, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2000-08-30 | | Release date: | 2000-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of N-acyl-D-glucosamine 2-epimerase from porcine kidney at 2.0 A resolution.
J.Mol.Biol., 303, 2000
|
|
8IY6
 
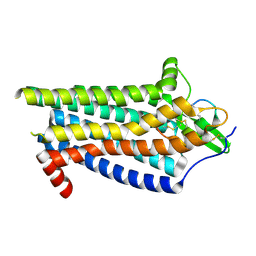 | | ETB-Gi complex bound to Endotheline-1, focused on receptor | | Descriptor: | Endothelin type B receptor, Endothelin-1 | | Authors: | Sano, F.K, Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-16 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of the endothelin-1-ET B -G i complex.
Elife, 12, 2023
|
|
8IY5
 
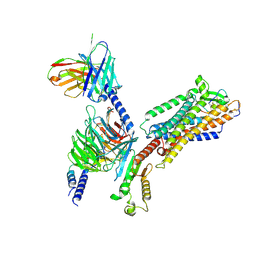 | | ETB-Gi complex bound to endothelin-1 | | Descriptor: | Endothelin type B receptor, Endothelin-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Sano, F.K, Akasaka, H, Shihoya, W, Nureki, O. | | Deposit date: | 2023-04-04 | | Release date: | 2023-08-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the endothelin-1-ET B -G i complex.
Elife, 12, 2023
|
|
7E4S
 
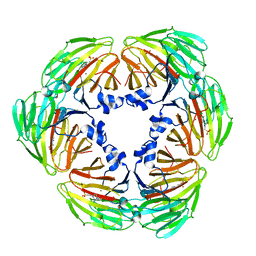 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-dehydro-4-deoxy-D-glucuronate isomerase, ZINC ION | | Authors: | Yamamoto, Y, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-02-15 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
7VGK
 
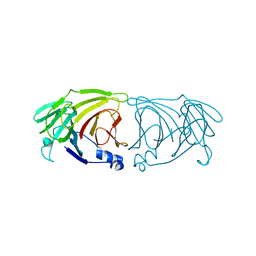 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI | | Descriptor: | 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase | | Authors: | Iwase, H, Oiki, S, Mikami, B, Takase, R, Hashimoto, W. | | Deposit date: | 2021-09-16 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of Lacticaseibacillus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI in complex with substrate analogs
J.Appl.Glyosci., 2023
|
|
7VEW
 
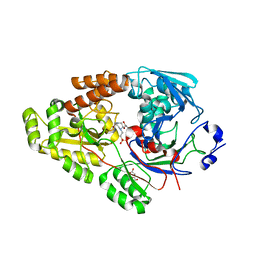 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with unsaturated trigalacturonic acid | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEQ
 
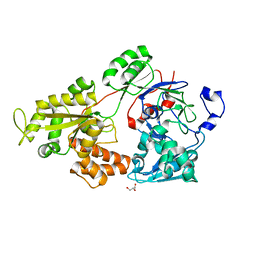 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in an open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VET
 
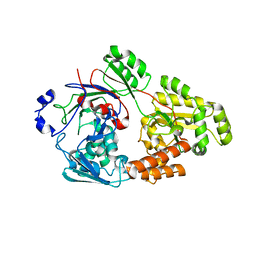 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a closed conformation | | Descriptor: | SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEV
 
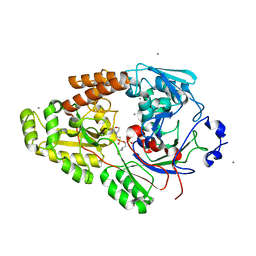 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VER
 
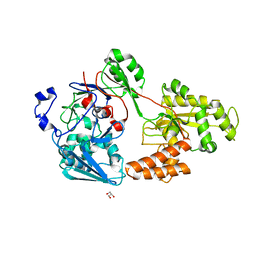 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in a full open conformation | | Descriptor: | GLYCEROL, SPH1118 | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
7VEU
 
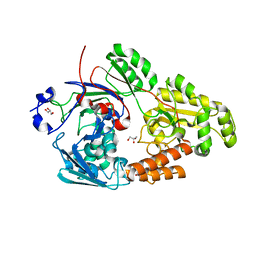 | | Crystal structure of bacterial chemotaxis-dependent pectin-binding protein SPH1118 in complex with galacturonic acid | | Descriptor: | GLYCEROL, SPH1118, alpha-D-galactopyranuronic acid | | Authors: | Anamizu, K, Takase, R, Hio, M, Watanebe, D, Mikami, B, Hashimoto, W. | | Deposit date: | 2021-09-10 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Substrate size-dependent conformational changes of bacterial pectin-binding protein crucial for chemotaxis and assimilation.
Sci Rep, 12, 2022
|
|
6JBO
 
 | | Crystal structure of EfeO-like protein Algp7 containing samarium ion | | Descriptor: | 1,2-ETHANEDIOL, Alginate-binding protein, CITRIC ACID | | Authors: | Okumura, K, Takase, R, Maruyama, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2019-01-26 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Rare metal binding by a cell-surface component of bacterial EfeUOB iron importer
To Be Published
|
|
6INZ
 
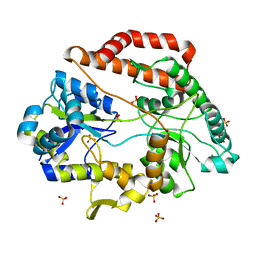 | | Crystal structure of solute-binding protein complexed with unsaturated hyaluronan disaccharide | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Extracellular solute-binding protein family 1, ... | | Authors: | Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.289 Å) | | Cite: | Substrate recognition by bacterial solute-binding protein is responsible for import of extracellular hyaluronan and chondroitin sulfate from the animal host.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
6K1Q
 
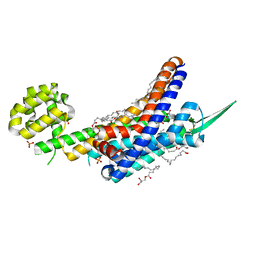 | | Human endothelin receptor type-B in complex with inverse agonist IRL2500 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-2-[[(2~{R})-2-[(3,5-dimethylphenyl)carbonyl-methyl-amino]-3-(4-phenylphenyl)propanoyl]amino]-3-(1~{H}-indol-3-yl)propanoic acid, Endothelin B receptor,Endolysin,Endothelin B receptor, ... | | Authors: | Nagiri, C, Shihoya, W, Nureki, O. | | Deposit date: | 2019-05-11 | | Release date: | 2019-07-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human endothelin ETBreceptor in complex with peptide inverse agonist IRL2500.
Commun Biol, 2, 2019
|
|
6JHX
 
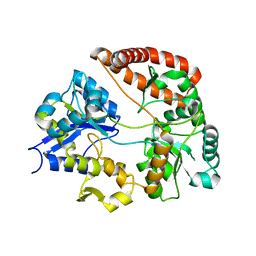 | |
2OKX
 
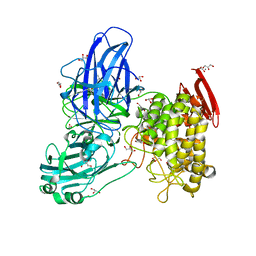 | | Crystal structure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A | | Descriptor: | CALCIUM ION, GLYCEROL, Rhamnosidase B | | Authors: | Cui, Z, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase Family 78 alpha-L-Rhamnosidase from Bacillus sp. GL1
J.Mol.Biol., 374, 2007
|
|
7D20
 
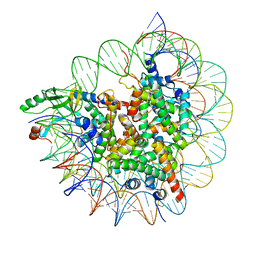 | | Cryo-EM structure of SET8-CENP-A-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
7D1Z
 
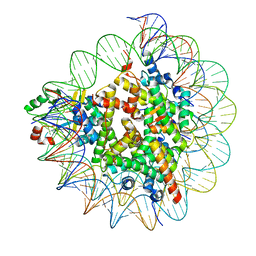 | | Cryo-EM structure of SET8-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ho, C.-H, Takizawa, Y, Kobayashi, W, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2020-09-15 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of nucleosomal histone H4 lysine 20 methylation by SET8 methyltransferase.
Life Sci Alliance, 4, 2021
|
|
