6AIS
 
 | |
6JP9
 
 | | Crsytal structure of a XMP complexed ATPPase subunit of M. jannaschii GMP synthetase | | Descriptor: | GMP synthase [glutamine-hydrolyzing] subunit B, MALONIC ACID, TRIETHYLENE GLYCOL, ... | | Authors: | Shivakumarasamy, S, Balaram, H. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic Insights into the Functioning of a Two-Subunit GMP Synthetase, an Allosterically Regulated, Ammonia Channeling Enzyme.
Biochemistry, 61, 2022
|
|
2RSV
 
 | |
6IEI
 
 | |
6ICS
 
 | |
6IDC
 
 | |
8HVB
 
 | |
8HVC
 
 | |
8HVD
 
 | |
8IE2
 
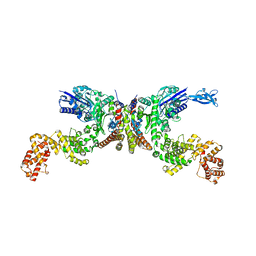 | | Crystal structure of Lactiplantibacillus plantarum GlyRS | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2023-02-15 | | Release date: | 2023-06-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Mechanism of tRNA recognition by heterotetrameric glycyl-tRNA synthetase from lactic acid bacteria.
J.Biochem., 174, 2023
|
|
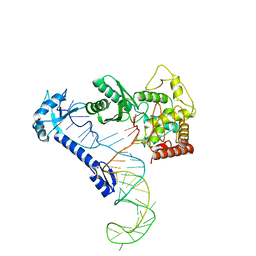
8IDF
 
 | |
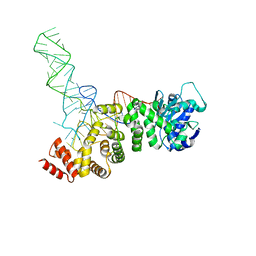
5HC9
 
 | |
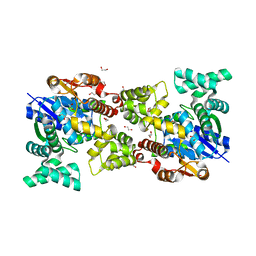
5Y0S
 
 | |
5Y0P
 
 | |
5Y0R
 
 | |
5Y0N
 
 | |
5Y0Q
 
 | | Crystal structure of BsTmcAL bound with AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, UPF0348 protein B4417_3650 | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis.
Nat. Chem. Biol., 14, 2018
|
|
5Y0O
 
 | | Crystal structure of apo BsTmcAL | | Descriptor: | UPF0348 protein B4417_3650 | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis.
Nat. Chem. Biol., 14, 2018
|
|
5Y0T
 
 | |
6K3I
 
 | |
5WU3
 
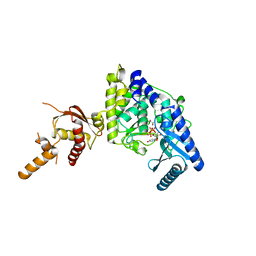 | | Crystal structure of human Tut1 bound with MgUTP, form II | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
5WU5
 
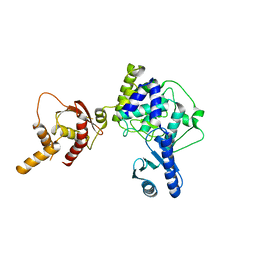 | | Crystal structure of apo human Tut1, form III | | Descriptor: | Speckle targeted PIP5K1A-regulated poly(A) polymerase | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2016-12-16 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Crystal structures of U6 snRNA-specific terminal uridylyltransferase
Nat Commun, 8, 2017
|
|
5WU2
 
 | |
5WU4
 
 | |
7FDD
 
 | | A Crystal structure of OspA mutant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Outer surface protein A | | Authors: | Shiga, S, Makabe, K. | | Deposit date: | 2021-07-16 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | beta-Strand-mediated Domain-swapping in the Absence of Hydrophobic Core Repacking.
J.Mol.Biol., 436, 2024
|
|
