2DA1
 
 | | Solution structure of the first homeobox domain of AT-binding transcription factor 1 (ATBF1) | | Descriptor: | Alpha-fetoprotein enhancer binding protein | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first homeobox domain of AT-binding transcription factor 1 (ATBF1)
To be Published
|
|
2DAD
 
 | | Solution structure of the fifth crystall domain of the non-lens protein, Absent in melanoma 1 | | Descriptor: | Absent in melanoma 1 protein | | Authors: | Ohnishi, S, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the fifth crystall domain of the non-lens protein, Absent in melanoma 1
To be Published
|
|
2DA7
 
 | | Solution structure of the homeobox domain of Zinc finger homeobox protein 1b (Smad interacting protein 1) | | Descriptor: | Zinc finger homeobox protein 1b | | Authors: | Ohnishi, S, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-12-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of Zinc finger homeobox protein 1b (Smad interacting protein 1)
To be Published
|
|
2DX6
 
 | | Crystal structure of conserved hypothetical protein, TTHA0132 from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, Hypothetical protein TTHA0132, ISOPROPYL ALCOHOL | | Authors: | Kishishita, S, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-24 | | Release date: | 2007-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of conserved hypothetical protein, TTHA0132 from Thermus thermophilus HB8
To be Published
|
|
2E29
 
 | | Solution structure of the GUCT domain from human ATP-dependent RNA helicase DDX50, DEAD box protein 50 | | Descriptor: | ATP-dependent RNA helicase DDX50 | | Authors: | Ohnishi, S, Paakkonen, K, Guntert, P, Sato, M, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-11-10 | | Release date: | 2007-05-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the GUCT domain from human RNA helicase II/Gubeta reveals the RRM fold, but implausible RNA interactions
Proteins, 74, 2008
|
|
2PH8
 
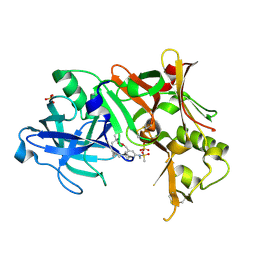 | | Crystal Structure of Human Beta Secretase Complexed with inhibitor | | Descriptor: | Beta-secretase 1, N-[(5R,14R)-5-AMINO-5,14-DIMETHYL-4-OXO-3-OXA-18-AZATRICYCLO[15.3.1.1~7,11~]DOCOSA-1(21),7(22),8,10,17,19-HEXAEN-19-YL]-N-METHYLMETHANESULFONAMIDE, SULFATE ION | | Authors: | Munshi, S. | | Deposit date: | 2007-04-10 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Strategies toward improving the brain penetration of macrocyclic tertiary carbinamine BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2CUF
 
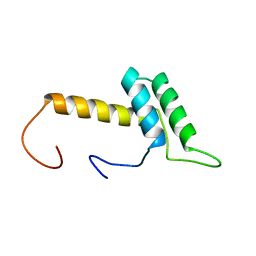 | | Solution structure of the homeobox domain of the human hypothetical protein FLJ21616 | | Descriptor: | FLJ21616 protein | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of the human hypothetical protein FLJ21616
To be Published
|
|
2CUE
 
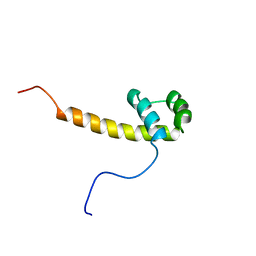 | | Solution structure of the homeobox domain of the human paired box protein Pax-6 | | Descriptor: | Paired box protein Pax6 | | Authors: | Ohnishi, S, Kigawa, T, Tochio, N, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of the human paired box protein Pax-6
To be Published
|
|
2CUB
 
 | | Solution structure of the SH3 domain of the human cytoplasmic protein Nck1 | | Descriptor: | Cytoplasmic protein NCK1 | | Authors: | Ohnishi, S, Kigawa, T, Sato, M, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-26 | | Release date: | 2005-11-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of the human cytoplasmic protein Nck1
To be Published
|
|
2DMV
 
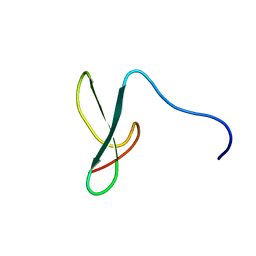 | | Solution structure of the second ww domain of Itchy homolog E3 ubiquitin protein ligase (Itch) | | Descriptor: | Itchy homolog E3 ubiquitin protein ligase | | Authors: | Ohnishi, S, Paakkonen, K, Tochio, N, Tomizawa, T, Koshiba, S, Inoue, M, Guntert, P, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second ww domain of Itchy homolog E3 ubiquitin protein ligase (Itch)
To be Published
|
|
2DMX
 
 | | Solution structure of the J domain of DnaJ homolog subfamily B member 8 | | Descriptor: | DnaJ homolog subfamily B member 8 | | Authors: | Ohnishi, S, Tochio, N, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the J domain of DnaJ homolog subfamily B member 8
To be Published
|
|
2DMW
 
 | | Solution structure of the LONGIN domain of Synaptobrevin-like protein 1 | | Descriptor: | synaptobrevin-like 1 variant | | Authors: | Ohnishi, S, Sato, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the LONGIN domain of Synaptobrevin-like protein 1
To be Published
|
|
2DWV
 
 | | Solution structure of the second WW domain from mouse salvador homolog 1 protein (mWW45) | | Descriptor: | Salvador homolog 1 protein | | Authors: | Ohnishi, S, Kigawa, T, Koshiba, S, Tomizawa, T, Sato, M, Tochio, N, Inoue, M, Harada, T, Watanabe, S, Guntert, P, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-17 | | Release date: | 2007-02-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an atypical WW domain in a novel beta-clam-like dimeric form
Febs Lett., 581, 2007
|
|
2DA6
 
 | | Solution structure of the homeobox domain of Hepatocyte nuclear factor 1-beta (HNF-1beta) | | Descriptor: | Hepatocyte nuclear factor 1-beta | | Authors: | Ohnishi, S, Kigawa, T, Sato, M, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-12-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of Hepatocyte nuclear factor 1-beta (HNF-1beta)
To be Published
|
|
2DA2
 
 | | Solution structure of the second homeobox domain of AT-binding transcription factor 1 (ATBF1) | | Descriptor: | Alpha-fetoprotein enhancer binding protein | | Authors: | Ohnishi, S, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second homeobox domain of AT-binding transcription factor 1 (ATBF1)
To be Published
|
|
2DA3
 
 | | Solution structure of the third homeobox domain of AT-binding transcription factor 1 (ATBF1) | | Descriptor: | Alpha-fetoprotein enhancer binding protein | | Authors: | Ohnishi, S, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third homeobox domain of AT-binding transcription factor 1 (ATBF1)
To be Published
|
|
2DA9
 
 | | Solution structure of the third SH3 domain of SH3-domain kinase binding protein 1 (Regulator of ubiquitous kinase, Ruk) | | Descriptor: | SH3-domain kinase binding protein 1 | | Authors: | Ohnishi, S, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-13 | | Release date: | 2006-06-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third SH3 domain of SH3-domain kinase binding protein 1 (Regulator of ubiquitous kinase, Ruk)
To be Published
|
|
2DMS
 
 | | Solution structure of the homeobox domain of Homeobox protein OTX2 | | Descriptor: | Homeobox protein OTX2 | | Authors: | Ohnishi, S, Saito, K, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-04-24 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain of Homeobox protein OTX2
To be Published
|
|
2DWC
 
 | | Crystal structure of Probable phosphoribosylglycinamide formyl transferase from Pyrococcus horikoshii OT3 complexed with ADP | | Descriptor: | 433aa long hypothetical phosphoribosylglycinamide formyl transferase, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION | | Authors: | Yoshikawa, S, Arai, R, Kamo-Uchikubo, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-10 | | Release date: | 2007-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Probable phosphoribosylglycinamide formyl transferase from Pyrococcus horikoshii OT3 complexed with ADP
To be Published
|
|
2E19
 
 | | Solution structure of the homeobox domain from human NIL-2-A zinc finger protein, transcription factor 8 | | Descriptor: | Transcription factor 8 | | Authors: | Ohnishi, S, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-19 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the homeobox domain from human NIL-2-A zinc finger protein, transcription factor 8
To be Published
|
|
3VOC
 
 | | Crystal structure of the catalytic domain of beta-amylase from paenibacillus polymyxa | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta/alpha-amylase, ... | | Authors: | Nishimura, S, Fujioka, T, Nakaniwa, T, Tada, T. | | Deposit date: | 2012-01-21 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural analysis by X-ray crystallography and small-angle scattering of the multi-domain beta-amylase from Paenibacillus polymyxa
To be Published
|
|
1V8L
 
 | | Structure Analysis of the ADP-ribose pyrophosphatase complexed with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-10 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8R
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase complexed with ADP-ribose and Zn | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2005-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8Y
 
 | | Crystal structure analysis of the ADP-ribose pyrophosphatase of E86Q mutant, complexed with ADP-ribose and Zn | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribose pyrophosphatase, ZINC ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
1V8D
 
 | | Crystal structure of the conserved hypothetical protein TT1679 from Thermus thermophilus | | Descriptor: | ZINC ION, hypothetical protein (TT1679) | | Authors: | Kishishita, S, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-05 | | Release date: | 2004-07-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of the conserved hypothetical protein TT1679 from Thermus thermophilus HB8
To be Published
|
|
