5ZWF
 
 | | Covalent bond formation between histidine of Vitamin D receptor (VDR) and a full agonist having a enone with a beta methyl group via conjugate addition reaction | | Descriptor: | (E,7R)-7-[(1R,3aS,4E,7aR)-7a-methyl-4-[2-[(3R,5R)-4-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3a,5,6,7-hexahydro-1H-inden-1-yl]oct-2-en-4-one, 13-meric peptide from DRIP205 NR2 BOX peptide, Vitamin D3 receptor | | Authors: | Yoshizawa, M, Itoh, T, Anami, Y, Kato, A, Yoshimoto, N, Yamamoto, K. | | Deposit date: | 2018-05-15 | | Release date: | 2018-07-18 | | Last modified: | 2018-08-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the Histidine Residue in Vitamin D Receptor That Covalently Binds to Electrophilic Ligands
J. Med. Chem., 61, 2018
|
|
1K3Z
 
 | | X-ray crystal structure of the IkBb/NF-kB p65 homodimer complex | | Descriptor: | Transcription factor p65, transcription factor inhibitor I-kappa-B-beta | | Authors: | Shiva, M, Huang, D.B, Chen, Y, Huxford, T, Ghosh, S, Ghosh, G. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of an IkappaBbeta x NF-kappaB p65 homodimer complex.
J.Biol.Chem., 278, 2003
|
|
2LMU
 
 | | Androcam at high calcium | | Descriptor: | CALCIUM ION, Calmodulin-related protein 97A | | Authors: | Joshi, M.K, Moran, S.T, Beckingham, K.M, Mackenzie, K.R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of androcam supports specialized interactions with myosin VI.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LMV
 
 | | Androcam at high calcium with three explicit Ca2+ | | Descriptor: | CALCIUM ION, Calmodulin-related protein 97A | | Authors: | Joshi, M.K, Moran, S.T, Beckingham, K.M, Mackenzie, K.R. | | Deposit date: | 2011-12-12 | | Release date: | 2012-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of androcam supports specialized interactions with myosin VI.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LM8
 
 | |
1X23
 
 | | Crystal structure of ubch5c | | Descriptor: | Ubiquitin-conjugating enzyme E2 D3 | | Authors: | Nakanishi, M, Teshima, N, Mizushima, T, Murata, S, Tanaka, K, Yamane, T. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of ubch5c
To be Published
|
|
5YJS
 
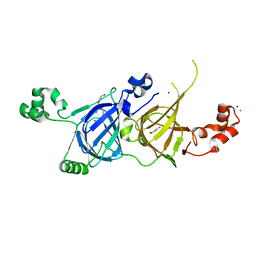 | | Structure of vicilin from Capsicum annuum | | Descriptor: | 2-HYDROXYBENZOIC ACID, CHLORIDE ION, COPPER (I) ION, ... | | Authors: | Shikhi, M, Nair, D.T, Salunke, D.M. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-guided identification of function: role ofCapsicum annuumvicilin during oxidative stress.
Biochem. J., 475, 2018
|
|
3VKW
 
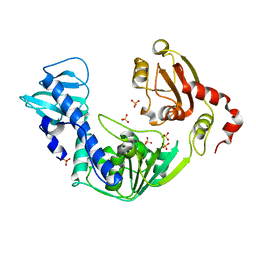 | | Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus | | Descriptor: | Replicase large subunit, SULFATE ION | | Authors: | Nishikiori, M, Sugiyama, S, Xiang, H, Niiyama, M, Ishibashi, K, Inoue, T, Ishikawa, M, Matsumura, H, Katoh, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superfamily 1 helicase from tomato mosaic virus
J.Virol., 86, 2012
|
|
1ULX
 
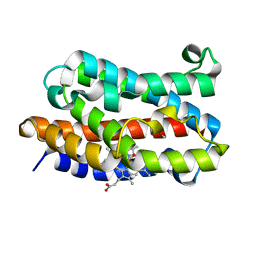 | | Partially photolyzed structure of CO-bound heme-heme oxygenase complex | | Descriptor: | CARBON MONOXIDE, Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-09-16 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CO-trapping site in heme oxygenase revealed by photolysis of its co-bound heme complex: mechanism of escaping from product inhibition
J.Mol.Biol., 341, 2004
|
|
1UGN
 
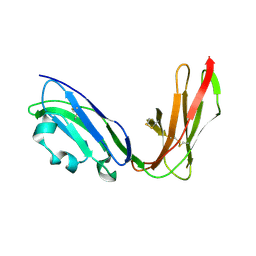 | | Crystal structure of LIR1.02, one of the alleles of LIR1 | | Descriptor: | Leukocyte immunoglobulin-like receptor 1 | | Authors: | Shiroishi, M, Rasubala, L, Kuroki, K, Amano, K, Tsuchiya, N, Tokunaga, K, Kohda, D, Maenaka, K. | | Deposit date: | 2003-06-17 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Extensive polymorphisms of LILRB1 (ILT2, LIR1) and their association with HLA-DRB1 shared epitope negative rheumatoid arthritis.
Hum.Mol.Genet., 14, 2005
|
|
1WOV
 
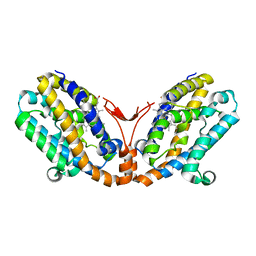 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
1WOW
 
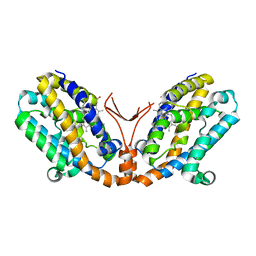 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 complexed with heme in ferrous form | | Descriptor: | Heme oxygenase 2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
5XI9
 
 | |
5XIR
 
 | |
1WE1
 
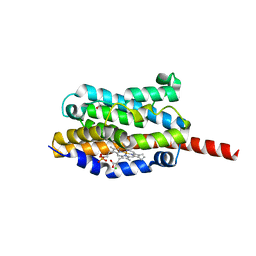 | | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC6803 in complex with heme | | Descriptor: | CHLORIDE ION, Heme oxygenase 1, ISOPROPYL ALCOHOL, ... | | Authors: | Sugishima, M, Migita, C.T, Zhang, X, Yoshida, T, Fukuyama, K. | | Deposit date: | 2004-05-21 | | Release date: | 2004-12-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of heme oxygenase-1 from cyanobacterium Synechocystis sp. PCC 6803 in complex with heme
Eur.J.Biochem., 271, 2004
|
|
1X60
 
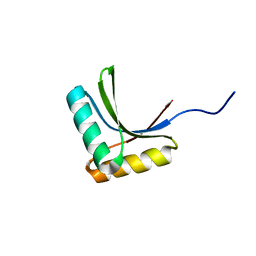 | | Solution structure of the peptidoglycan binding domain of B. subtilis cell wall lytic enzyme CwlC | | Descriptor: | Sporulation-specific N-acetylmuramoyl-L-alanine amidase | | Authors: | Mishima, M, Shida, T, Yabuki, K, Kato, K, Sekiguchi, J, Kojima, C. | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Peptidoglycan Binding Domain of Bacillus subtilis Cell Wall Lytic Enzyme CwlC: Characterization of the Sporulation-Related Repeats by NMR(,)
Biochemistry, 44, 2005
|
|
5YO8
 
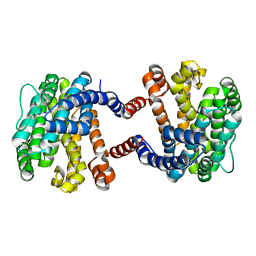 | |
1WOX
 
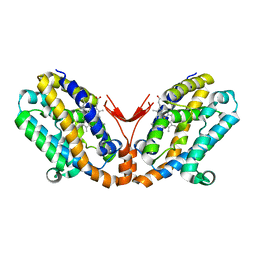 | | Crystal structure of heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme and NO | | Descriptor: | Heme oxygenase 2, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Hagiwara, Y, Zhang, X, Yoshida, T, Migita, C.T, Fukuyama, K. | | Deposit date: | 2004-08-26 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of dimeric heme oxygenase-2 from Synechocystis sp. PCC 6803 in complex with heme.
Biochemistry, 44, 2005
|
|
1UGU
 
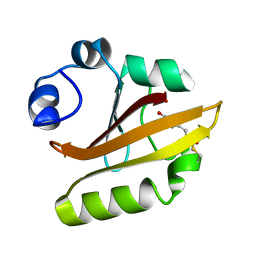 | | Crystal structure of PYP E46Q mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Sugishima, M, Tanimoto, Y, Hamada, N, Tokunaga, F, Fukuyama, K. | | Deposit date: | 2003-06-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of photoactive yellow protein (PYP) E46Q mutant at 1.2 A resolution suggests how Glu46 controls the spectroscopic and kinetic characteristics of PYP.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UFU
 
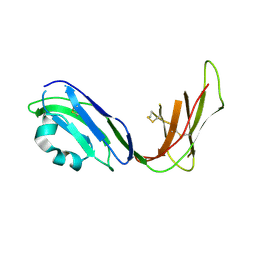 | | Crystal structure of ligand binding domain of immunoglobulin-like transcript 2 (ILT2; LIR-1) | | Descriptor: | Immunoglobulin-like transcript 2 | | Authors: | Shiroishi, M, Amano, K, Rasubala, L, Tsumoto, K, Kumagai, I, Kohda, D, Maenaka, K. | | Deposit date: | 2003-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetic and thermodynamic properties of the interaction between Immunoglobulin like transcript (ILT) and MHC class I
To be Published
|
|
3VTB
 
 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 25-adamantyl-23-yne-19-norvitammin D ADTK1 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6S)-6-hydroxy-6-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hex-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
3VTC
 
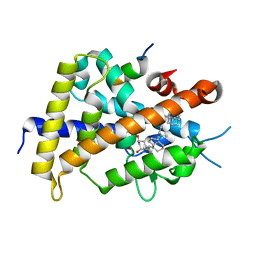 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 26-adamantyl-23-yne-19-norvitammin D ADTK3 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6R)-6-hydroxy-7-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hept-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, 1,2-ETHANEDIOL, COACTIVATOR PEPTIDE DRIP, ... | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
3VTD
 
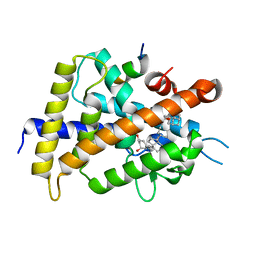 | | Crystal structure of rat vitamin D receptor bound to a partial agonist 26-adamantyl-23-yne-19-norvitammin D ADTK4 | | Descriptor: | (1R,3R,7E,17beta)-17-{(2R,6S)-6-hydroxy-7-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-yl]hept-4-yn-2-yl}-2-methylidene-9,10-secoestra-5,7-diene-1,3-diol, COACTIVATOR PEPTIDE DRIP, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Kudo, T, Tokiwa, H, Makishima, M, Yamada, S, Ikura, T, Ito, N. | | Deposit date: | 2012-05-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Combination of Triple Bond and Adamantane Ring on the Vitamin D Side Chain Produced Partial Agonists for Vitamin D Receptor.
J.Med.Chem., 2014
|
|
3WDP
 
 | | Structural analysis of a beta-glucosidase mutant derived from a hyperthermophilic tetrameric structure | | Descriptor: | Beta-glucosidase, GLYCEROL, PHOSPHATE ION | | Authors: | Nakabayashi, M, Kataoka, M, Ishikawa, K. | | Deposit date: | 2013-06-19 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of beta-glucosidase mutants derived from a hyperthermophilic tetrameric structure.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3WQ8
 
 | | Monomer structure of hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form | | Descriptor: | Beta-glucosidase | | Authors: | Nakabayashi, M, Kataoka, M, Watanabe, M, Ishikawa, K. | | Deposit date: | 2014-01-23 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Monomer structure of a hyperthermophilic beta-glucosidase mutant forming a dodecameric structure in the crystal form.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
