2ELD
 
 | |
2ED3
 
 | |
2EMU
 
 | |
2ENU
 
 | |
2EMR
 
 | |
2EN5
 
 | |
2E4N
 
 | |
2EH4
 
 | |
2ED5
 
 | |
2ELE
 
 | |
2EH5
 
 | |
1V7Y
 
 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli at room temperature | | Descriptor: | SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-25 | | Release date: | 2005-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
1WLF
 
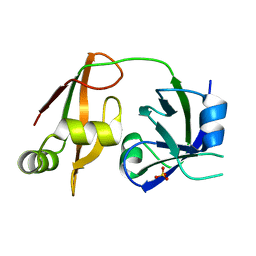 | | Structure of the N-terminal domain of PEX1 AAA-ATPase: Characterization of a putative adaptor-binding domain | | Descriptor: | Peroxisome biogenesis factor 1, SULFATE ION | | Authors: | Shiozawa, K, Maita, N, Tomii, K, Seto, A, Goda, N, Tochio, H, Akiyama, Y, Shimizu, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the N-terminal Domain of PEX1 AAA-ATPase: CHARACTERIZATION OF A PUTATIVE ADAPTOR-BINDING DOMAIN
J.Biol.Chem., 279, 2004
|
|
1WQ5
 
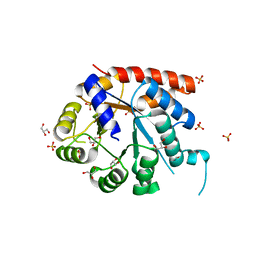 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli | | Descriptor: | GLYCEROL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-22 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
2ZPG
 
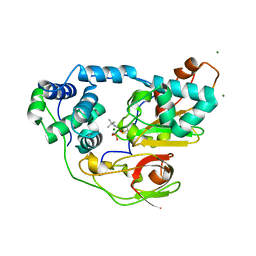 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 120min at 293K | | Descriptor: | FE (III) ION, MAGNESIUM ION, Nitrile hydratase subunit alpha, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPE
 
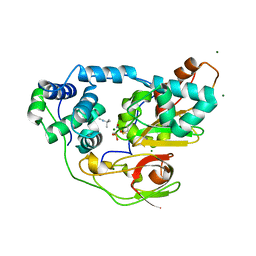 | | nitrosylated Fe-type nitrile hydratase with tert-butylisonitrile | | Descriptor: | FE (III) ION, MAGNESIUM ION, NITRIC OXIDE, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPF
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 18min at 293K | | Descriptor: | FE (III) ION, MAGNESIUM ION, NITRIC OXIDE, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPI
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 440min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.491 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPB
 
 | | nitrosylated Fe-type nitrile hydratase | | Descriptor: | FE (III) ION, MAGNESIUM ION, NITRIC OXIDE, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-09 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZPH
 
 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 340min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
2ZV3
 
 | |
1V8G
 
 | |
1X1O
 
 | |
1WN2
 
 | |
1WU8
 
 | |
