5X7E
 
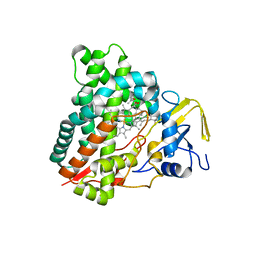 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R84A mutant) in complex with 1,25-dihydroxyvitamin D2 | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(E,2R,5S)-5,6-dimethyl-6-oxidanyl-hept-3-en-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydr o-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, PROTOPORPHYRIN IX CONTAINING FE, Vitamin D3 dihydroxylase | | Authors: | Hayashi, K, Yasuda, K, Shiro, Y, Sugimoto, H, Sakaki, T. | | Deposit date: | 2017-02-25 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Production of an active form of vitamin D2 by genetically engineered CYP105A1
Biochem. Biophys. Res. Commun., 486, 2017
|
|
2D29
 
 | |
2D3K
 
 | |
3AGK
 
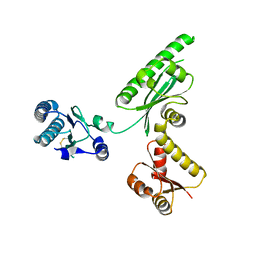 | | Crystal structure of archaeal translation termination factor, aRF1 | | Descriptor: | Peptide chain release factor subunit 1 | | Authors: | Kobayashi, K, Kikuno, I, Ishitani, R, Ito, K, Nureki, O. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Omnipotent role of archaeal elongation factor 1 alpha (EF1{alpha}) in translational elongation and termination, and quality control of protein synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AV4
 
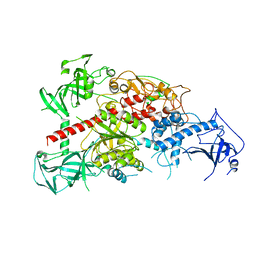 | | Crystal structure of mouse DNA methyltransferase 1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV6
 
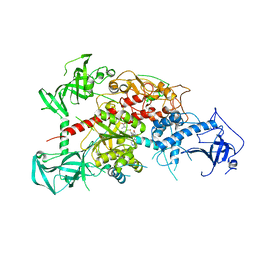 | | Crystal structure of mouse DNA methyltransferase 1 with AdoMet | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV5
 
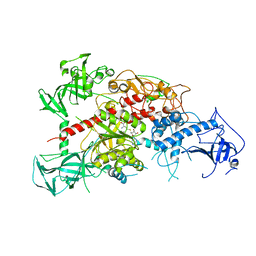 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1WQB
 
 | |
5Y4N
 
 | | Crystal structure of aerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J.Inorg.Biochem., 177, 2017
|
|
5XLG
 
 | | Crystal structure of anaerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLE
 
 | | Crystal structure of anaerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
5XLF
 
 | | Crystal structure of aerobically purified and aerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
1V8G
 
 | |
5XLH
 
 | | Crystal structure of aerobically purified and aerobically crystallized for 12weeks D. vulgaris Miyazaki F [NiFe]-hydrogenase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | Deposit date: | 2017-05-10 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J. Inorg. Biochem., 177, 2017
|
|
1RIL
 
 | | CRYSTAL STRUCTURE OF RIBONUCLEASE H FROM THERMUS THERMOPHILUS HB8 REFINED AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Okumura, M, Katayanagi, K, Kimura, S, Kanaya, S, Nakamura, H, Morikawa, K. | | Deposit date: | 1993-01-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 A resolution.
J.Mol.Biol., 230, 1993
|
|
1RBS
 
 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
1RBU
 
 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
5XTM
 
 | | Crystal structure of PhoRpp38 bound to a K-turn in P12.2 helix | | Descriptor: | 50S ribosomal protein L7Ae, MAGNESIUM ION, RNA (47-MER) | | Authors: | Oshima, K, Kimura, M. | | Deposit date: | 2017-06-20 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the archaeal RNase P protein Rpp38 in complex with RNA fragments containing a K-turn motif.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1RBV
 
 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
1RBR
 
 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
1RBT
 
 | | STRUCTURAL STUDY OF MUTANTS OF ESCHERICHIA COLI RIBONUCLEASE HI WITH ENHANCED THERMOSTABILITY | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Kimura, S, Kanaya, S, Morikawa, K, Nakamura, H. | | Deposit date: | 1993-02-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study of mutants of Escherichia coli ribonuclease HI with enhanced thermostability.
Protein Eng., 6, 1993
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
5Y7M
 
 | | Crystal structure of PhoRpp38 bound to a K-turn in P12.1 helix | | Descriptor: | 50S ribosomal protein L7Ae, GUANOSINE-5'-TRIPHOSPHATE, RNA (52-MER), ... | | Authors: | Oshima, K, Kimura, M. | | Deposit date: | 2017-08-01 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the archaeal RNase P protein Rpp38 in complex with RNA fragments containing a K-turn motif.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1WLF
 
 | | Structure of the N-terminal domain of PEX1 AAA-ATPase: Characterization of a putative adaptor-binding domain | | Descriptor: | Peroxisome biogenesis factor 1, SULFATE ION | | Authors: | Shiozawa, K, Maita, N, Tomii, K, Seto, A, Goda, N, Tochio, H, Akiyama, Y, Shimizu, T, Shirakawa, M, Hiroaki, H. | | Deposit date: | 2004-06-25 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the N-terminal Domain of PEX1 AAA-ATPase: CHARACTERIZATION OF A PUTATIVE ADAPTOR-BINDING DOMAIN
J.Biol.Chem., 279, 2004
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
