6CLP
 
 | | 1.16 A MicroED structure of GSNQNNF at 2.5 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.16 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLB
 
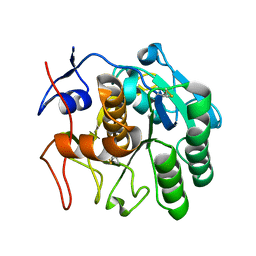 | | 3.20 A MicroED structure of proteinase K at 7.8 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.2 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLL
 
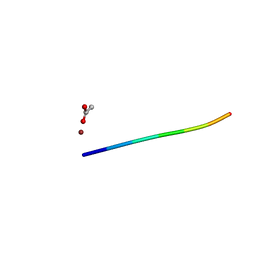 | | 1.02 A MicroED structure of GSNQNNF at 1.2 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.02 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLT
 
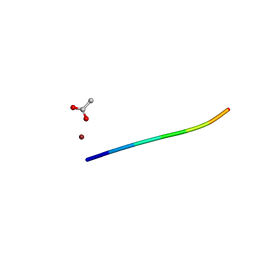 | | 1.45 A MicroED structure of GSNQNNF at 3.8 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.45 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLD
 
 | | 1.01 A MicroED structure of GSNQNNF at 0.81 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLK
 
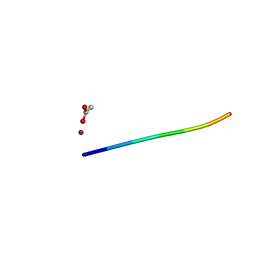 | | 1.01 A MicroED structure of GSNQNNF at 0.82 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLS
 
 | | 1.46 A MicroED structure of GSNQNNF at 3.4 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.46 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLF
 
 | | 1.15 A MicroED structure of GSNQNNF at 1.9 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLO
 
 | | 1.15 A MicroED structure of GSNQNNF at 2.1 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.15 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CL7
 
 | | 1.71 A MicroED structure of proteinase K at 0.86 e- / A^2 | | Descriptor: | Proteinase K | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.71 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLE
 
 | | 1.01 A MicroED structure of GSNQNNF at 1.3 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
6CLM
 
 | | 1.01 A MicroED structure of GSNQNNF at 1.5 e- / A^2 | | Descriptor: | ACETATE ION, GSNQNNF, ZINC ION | | Authors: | Hattne, J, Shi, D, Glynn, C, Zee, C.-T, Gallagher-Jones, M, Martynowycz, M.W, Rodriguez, J.A, Gonen, T. | | Deposit date: | 2018-03-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.01 Å) | | Cite: | Analysis of Global and Site-Specific Radiation Damage in Cryo-EM.
Structure, 26, 2018
|
|
3Q98
 
 | | Structure of ygeW encoded protein from E. coli | | Descriptor: | transcarbamylase | | Authors: | Li, Y, Jing, Z, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2011-01-07 | | Release date: | 2011-05-04 | | Last modified: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The ygeW encoded protein from Escherichia coli is a knotted ancestral catabolic transcarbamylase.
Proteins, 79, 2011
|
|
4KZT
 
 | | Structure mmNAGS bound with L-arginine | | Descriptor: | ARGININE, DI(HYDROXYETHYL)ETHER, N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Zhao, G, Jin, Z, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2013-05-30 | | Release date: | 2014-04-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of N-acetyl-L-glutamate synthase/kinase from Maricaulis maris with the allosteric inhibitor L-arginine bound.
Biochem.Biophys.Res.Commun., 437, 2013
|
|
3PEQ
 
 | | PPARd complexed with a phenoxyacetic acid partial agonist | | Descriptor: | IODIDE ION, Peroxisome proliferator-activated receptor delta, [(4-{butyl[2-methyl-4'-(methylsulfanyl)biphenyl-3-yl]sulfamoyl}naphthalen-1-yl)oxy]acetic acid, ... | | Authors: | Lambert, M.H, Evans, K.A, Shearer, B.G, Wisnoski, D.D, Shi, D, Jin, J, Rivero, R.A, Sparks, S.M, Winegar, D.A, Billin, A.N, Britt, C, Way, J.M, Leesnitzer, L.M, Merrihew, R.V. | | Deposit date: | 2010-10-27 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phenoxyacetic acid PPARd partial agonists for the treatment of type 2 diabetes: synthesis, optimization, and in vivo efficacy
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7K3Z
 
 | | P. falciparum Cpn60 D474A mutant bound to ATP | | Descriptor: | 60 kDa chaperonin, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Tolia, N.H, Shi, D, Nguyen, B. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.69 Å) | | Cite: | Crystal structure of P. falciparum Cpn60 bound to ATP reveals an open dynamic conformation before substrate binding.
Sci Rep, 11, 2021
|
|
