7DRG
 
 | | Pseudomonas aeruginosa T6SS protein PA0821 | | Descriptor: | PA0821 | | Authors: | She, Z. | | Deposit date: | 2020-12-28 | | Release date: | 2021-06-16 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Pseudomonas aeruginosa PAAR2 cluster encodes a putative VRR-NUC domain-containing effector.
Febs J., 288, 2021
|
|
7DYM
 
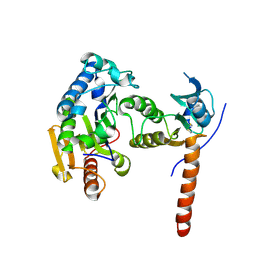 | | Pseudomonas aeruginosa TseT-TsiT complex | | Descriptor: | Imm52 domain-containing protein, Tox-REase-5 domain-containing protein | | Authors: | She, Z. | | Deposit date: | 2021-01-22 | | Release date: | 2021-08-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure and SAXS studies unveiled a novel inhibition mechanism of the Pseudomonas aeruginosa T6SS TseT-TsiT complex.
Int.J.Biol.Macromol., 188, 2021
|
|
6JDP
 
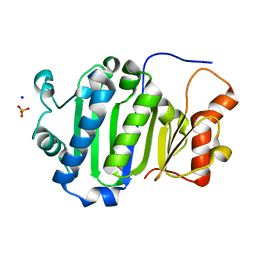 | |
6JQ4
 
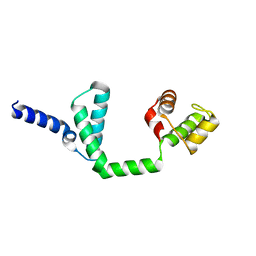 | | HIGA Escherichia coli-K12 | | Descriptor: | Antitoxin HigA | | Authors: | She, Z, Xu, B.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes of antitoxin HigA from Escherichia coli str. K-12 upon binding of its cognate toxin HigB reveal a new regulation mechanism in toxin-antitoxin systems.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
7FH4
 
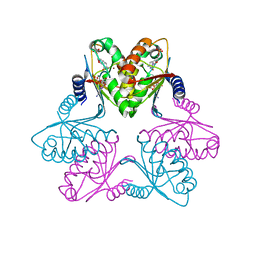 | | Chlorovirus PBCV-1 bi-functional dCMP/dCTP deaminase bi-DCD | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CMP/dCMP-type deaminase domain-containing protein, ... | | Authors: | She, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of a multi-functional deaminase in chlorovirus PBCV-1.
Arch.Biochem.Biophys., 727, 2022
|
|
6LCH
 
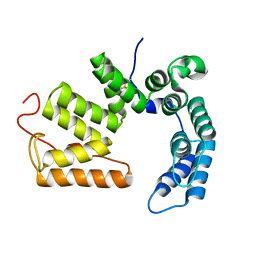 | | Pseudomonas aeruginosa 5086 (PA5086) | | Descriptor: | Sel1 repeat family protein | | Authors: | She, Z, Geng, Z. | | Deposit date: | 2019-11-19 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Characterization of the Pseudomonas aeruginosa T6SS PldB immunity proteins PA5086, PA5087 and PA5088 explains a novel stockpiling mechanism.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7FH9
 
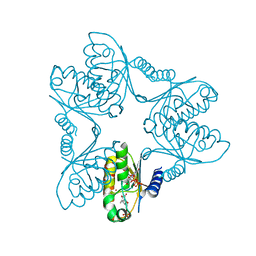 | |
5XMG
 
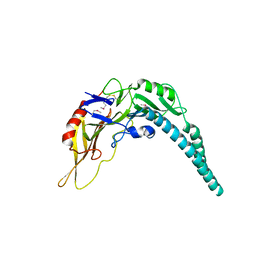 | | Crystal structure of PA3488 from Pseudomonas aeruginosa | | Descriptor: | Uncharacterized protein | | Authors: | She, Z, Gao, Z.Q. | | Deposit date: | 2017-05-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and SAXS analysis of Tle5-Tli5 complex reveals a novel inhibition mechanism of H2-T6SS in Pseudomonas aeruginosa.
Protein Sci., 26, 2017
|
|
4EQ6
 
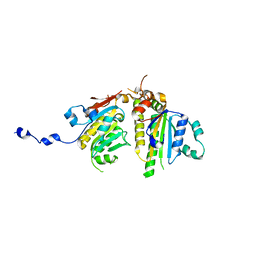 | |
8UAE
 
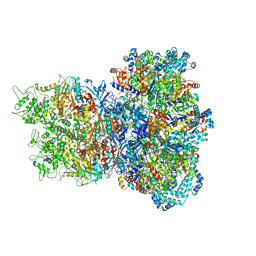 | | E. coli Sir2_HerA complex (12:6) with ATPgamaS | | Descriptor: | MAGNESIUM ION, Nucleoside triphosphate hydrolase, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
8UAF
 
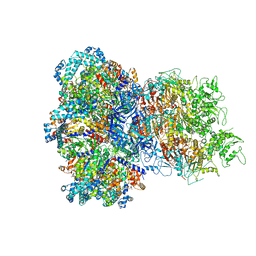 | | E. coli Sir2_HerA complex (12:6) bound with NAD+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Shen, Z.F, Lin, Q.P, Fu, T.M. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Assembly-mediated activation of the SIR2-HerA supramolecular complex for anti-phage defense.
Mol.Cell, 83, 2023
|
|
7WLH
 
 | |
5XBC
 
 | |
6LP9
 
 | | the protein of cat virus | | Descriptor: | nsp1 protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural and Biological Basis of Alphacoronavirus nsp1 Associated with Host Proliferation and Immune Evasion.
Viruses, 12, 2020
|
|
7VWV
 
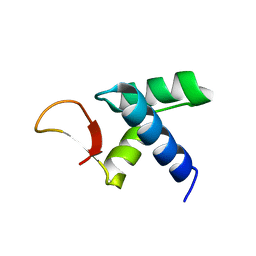 | |
6IVC
 
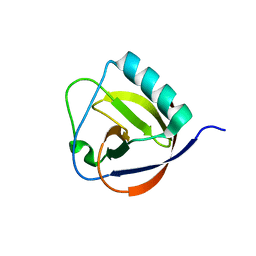 | | The full length of TGEV nsp1 | | Descriptor: | nsp1 protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
J.Biol.Chem., 294, 2019
|
|
6IVD
 
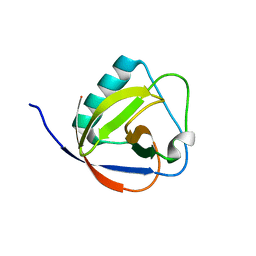 | | TGEV nsp1 mutant - 91-95sg | | Descriptor: | nsp1 mutant protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2018-12-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence.
J.Biol.Chem., 294, 2019
|
|
6LPA
 
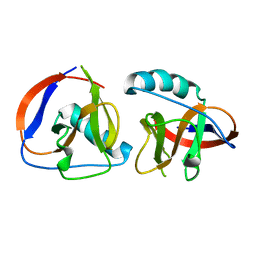 | | The nsp1 protein of a new porcine coronavirus | | Descriptor: | sp1 protein | | Authors: | Shen, Z, Peng, G.Q. | | Deposit date: | 2020-01-09 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural and Biological Basis of Alphacoronavirus nsp1 Associated with Host Proliferation and Immune Evasion.
Viruses, 12, 2020
|
|
1IH9
 
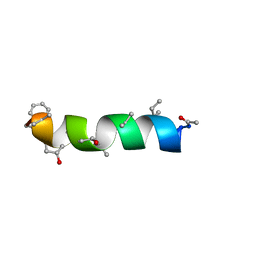 | | NMR Structure of Zervamicin IIB (peptaibol antibiotic) Bound to DPC Micelles | | Descriptor: | ZERVAMICIN IIB | | Authors: | Shenkarev, Z.O, Balasheva, T.A, Efremov, R.G, Yakimenko, Z.A, Ovchinnikova, T.V, Raap, J, Arseniev, A.S. | | Deposit date: | 2001-04-19 | | Release date: | 2002-02-13 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of Zervamicin Iib Bound to Dpc Micelles: Implications for Voltage-Gating.
Biophys.J., 82, 2002
|
|
7CQH
 
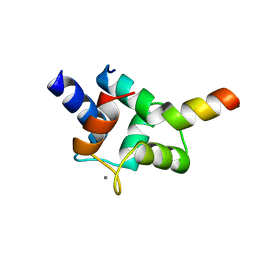 | | Complex of TRP_CBS2 and Calmodulin_Clobe | | Descriptor: | AT15141p, CALCIUM ION, Transient receptor potential protein | | Authors: | Shen, Z.S. | | Deposit date: | 2020-08-10 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Calmodulin binds to Drosophila TRP with an unexpected mode.
Structure, 29, 2021
|
|
7CQP
 
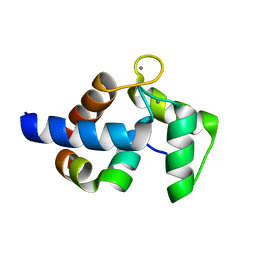 | | Complex of TRPC4 and Calmodulin_Nlobe | | Descriptor: | CALCIUM ION, Calmodulin-1, Peptide from Short transient receptor potential channel 4 | | Authors: | Shen, Z.S. | | Deposit date: | 2020-08-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calmodulin binds to Drosophila TRP with an unexpected mode.
Structure, 29, 2021
|
|
7CQV
 
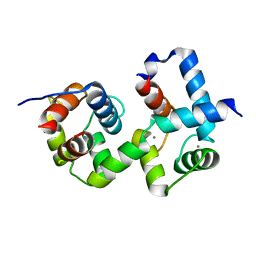 | | Complex of TRP_CBS1 and Calmodulin_Nlobe | | Descriptor: | AT15141p, CALCIUM ION, Transient receptor potential protein | | Authors: | Shen, Z.S. | | Deposit date: | 2020-08-11 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Calmodulin binds to Drosophila TRP with an unexpected mode.
Structure, 29, 2021
|
|
8EFV
 
 | |
1TE3
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TEQ
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, HYDROXIDE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-25 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
