7XU8
 
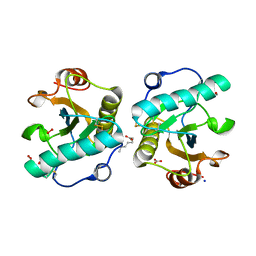 | | Structure of the complex of camel peptidoglycan recognition protein-short (PGRP-S) with heptanoic acid at 2.15 A resolution. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CARBONATE ION, ... | | Authors: | Maurya, A, Ahmad, N, Viswanathan, V, Singh, P.K, Yamini, S, Sharma, P, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-05-18 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand recognition by peptidoglycan recognition protein-S (PGRP-S): structure of the complex of camel PGRP-S with heptanoic acid at 2.15 angstrom resolution.
Int J Biochem Mol Biol, 13, 2022
|
|
5XGY
 
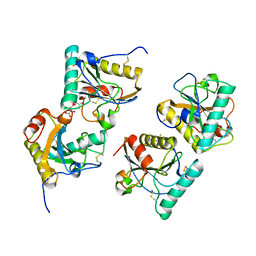 | | Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Shokeen, A, Sharma, P, Singh, P.K, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-04-18 | | Release date: | 2017-05-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of peptidoglycan recognition protein (PGRP-S) at 2.45 A resolution
To Be Published
|
|
3T39
 
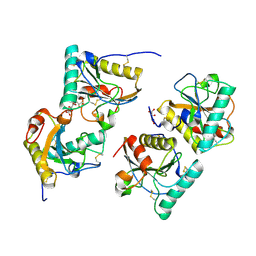 | | Crystal structure of the complex of camel peptidoglycan recognition protein(CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution | | Descriptor: | (3R,4S,5R)-3,4,5-TRIHYDROXYCYCLOHEX-1-ENE-1-CARBOXYLIC ACID, GLYCEROL, Peptidoglycan recognition protein 1, ... | | Authors: | Sharma, P, Dube, D, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-07-25 | | Release date: | 2011-08-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex of peptidoglycan recognition protein-short (CPGRP-S) with a mycobacterium metabolite shikimate at 2.7 A resolution
To be Published
|
|
8ZN1
 
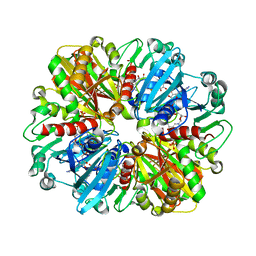 | | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution
To Be Published
|
|
8ZN4
 
 | | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution
To Be Published
|
|
8ZOZ
 
 | | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pahuja, P, Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the complex of glyceraldehyde-3-phosphate dehydrogenase of type B from Acinetobacter baumannii with Adenosine monophosphate at 3.20 A resolution.
To Be Published
|
|
6IDM
 
 | | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Bairagya, H.R, Shokeen, A, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution
To Be Published
|
|
6J93
 
 | | Crystal structure of Peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.95 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Iqbal, N, Sharma, S, Singh, T.P. | | Deposit date: | 2019-01-21 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Crystal structure of Peptidyl-tRNA hydrolase form apo at 0.95 A resolution.
To Be Published
|
|
6JQT
 
 | |
6JGU
 
 | |
6JJ1
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.97 A resolution with disordered five N-terminal residues | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase | | Authors: | Iqbal, N, Sharma, P, Chaudhary, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-24 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.97 A resolution with disordered five N-terminal residues
To Be Published
|
|
7WP3
 
 | | Crystal structure of the complex of proliferating cell nuclear antigen (PCNA) from Leishmania donovani with 1,5-Bis (4-amidinophenoxy) pentane (PNT) at 2.95 A resolution | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, Proliferating cell nuclear antigen | | Authors: | Ahmad, M.I, Yadav, S.P, Singh, P.K, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-01-22 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Crystal structure of the complex of proliferating cell nuclear antigen (PCNA) from Leishmania donovani with 1,5-Bis (4-amidinophenoxy) pentane (PNT) at 2.95 A resolution
To Be Published
|
|
5H0T
 
 | | Crystal structure of proliferating cell nuclear antigen from Leishmania donovani at 2.73 Angstrom resolution | | Descriptor: | DI(HYDROXYETHYL)ETHER, Proliferating cell nuclear antigen | | Authors: | Singh, P.K, Yadav, S.P, Sharma, P.S, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-10-06 | | Release date: | 2016-11-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure and binding studies of proliferating cell nuclear antigen from Leishmania donovani.
Biochim. Biophys. Acta, 1865, 2017
|
|
6JJQ
 
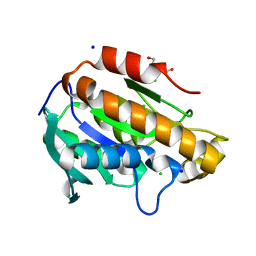 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Bairagya, H.R, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution.
To Be Published
|
|
7WZY
 
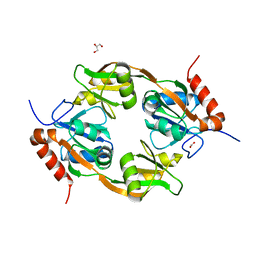 | | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 2.975 A resolution | | Descriptor: | ATP phosphoribosyltransferase, FORMIC ACID, GLYCEROL | | Authors: | Ahmad, N, Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-19 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.975 Å) | | Cite: | Crystal structure of Adenosine triphosphate phosphoribosyltransferase (HisG) from Acinetobacter baumannii at 2.975 A resolution
To Be Published
|
|
7XFW
 
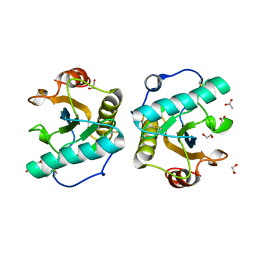 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.07 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure of the complex of camel peptidoglycan recognition protein-S with hexanoic acid reveals novel features of the versatile ligand-binding site at the dimeric interface.
Biochim Biophys Acta Proteins Proteom, 1871, 2022
|
|
7XFX
 
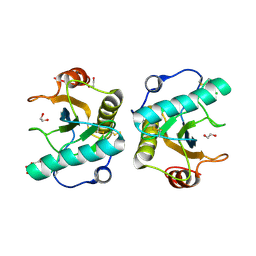 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.28 A resolution.
To Be Published
|
|
7XFY
 
 | | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Maurya, A, Singh, P.K, Viswanathan, V, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-04-02 | | Release date: | 2022-05-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of the ternary complex of Peptidoglycan recognition protein, PGRP-S with hexanoic and tartaric acids at 2.67 A resolution.
To Be Published
|
|
6KL8
 
 | | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of Piptidyl t-RNA hydrolase from Acinetobacter baumannii with bound NaCl at the substrate binding site
To Be Published
|
|
7WYJ
 
 | | Structure of the complex of lactoperoxidase with nitric oxide catalytic product nitrite at 1.89 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Pandey, N, Singh, A.K, Sinha, M, Singh, R.P, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural evidence of the conversion of nitric oxide (NO) to nitrite ion (NO2-) by lactoperoxidase (LPO): Structure of the complex of LPO with NO2- at 1.89 angstrom resolution
J.Inorg.Biochem., 247, 2023
|
|
6L2J
 
 | | Crystal structure of yak lactoperoxidase at 1.93 A resolution. | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-10-04 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 1.93 A resolution.
To Be Published
|
|
6KY7
 
 | | Crystal structure of yak lactoperoxidase at 2.27 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, P.K, Viswanathan, V, Sharma, P, Rani, C, Sharma, S, Singh, T.P. | | Deposit date: | 2019-09-16 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of yak lactoperoxidase at 2.27 A resolution
To Be Published
|
|
6LSP
 
 | |
7VE3
 
 | | Structure of the complex of sheep lactoperoxidase with hypoiodite at 2.70 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, IODIDE ION, ... | | Authors: | Singh, P.K, Yamini, S, Singh, R.P, Singh, A.K, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-09-07 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence of the oxidation of iodide ion into hyper-reactive hypoiodite ion by mammalian heme lactoperoxidase.
Protein Sci., 31, 2022
|
|
6JKX
 
 | | Crystal structure of peptidyl-tRNA hydrolase with multiple sodium and chloride ions at 1.08 A resolution. | | Descriptor: | CHLORIDE ION, METHANOL, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Sharma, P, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2019-03-03 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase with multiple sodium and chloride ions at 1.08 A resolution.
To Be Published
|
|
