2XDJ
 
 | |
5C5V
 
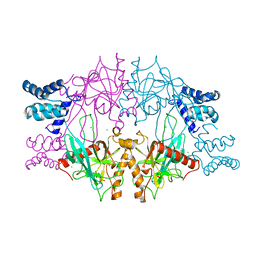 | | Recombinant Inorganic Pyrophosphatase from T brucei brucei | | Descriptor: | 1,2-ETHANEDIOL, Acidocalcisomal pyrophosphatase, BROMIDE ION, ... | | Authors: | Jamwal, A, Yogavel, M, Sharma, A. | | Deposit date: | 2015-06-22 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and Functional Highlights of Vacuolar Soluble Protein 1 from Pathogen Trypanosoma brucei brucei
J.Biol.Chem., 290, 2015
|
|
4Q45
 
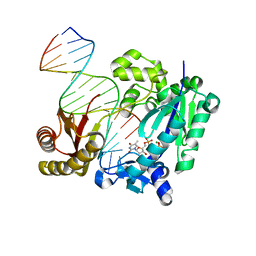 | | DNA Polymerase- damaged DNA complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*TP*CP*TP*A*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*CP*TP*AP*GP*GP*(RDG)P*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Kottur, J, Sharma, A, Nair, D.T. | | Deposit date: | 2014-04-13 | | Release date: | 2015-05-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.176 Å) | | Cite: | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
4DEZ
 
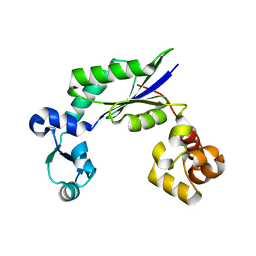 | | Structure of MsDpo4 | | Descriptor: | DNA polymerase IV 1 | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2012-01-22 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The PAD region in the mycobacterial DinB homologue MsPolIV exhibits positional heterogeneity.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6A88
 
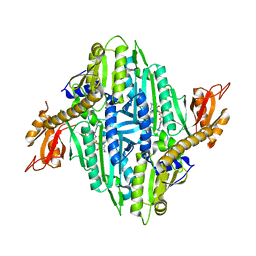 | | Crystal Structure of T. gondii prolyl tRNA synthetase with Febrifugine and ATP Analog | | Descriptor: | 3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Kumari, S, Mishra, S, Yogavel, M, Sharma, A. | | Deposit date: | 2018-07-06 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Conformational heterogeneity in apo and drug-bound structures of Toxoplasma gondii prolyl-tRNA synthetase.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4Q44
 
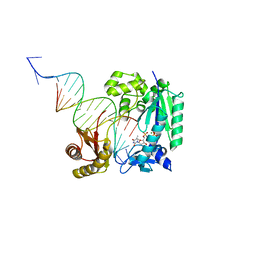 | | Polymerase-Damaged DNA Complex | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*TP*CP*TP*AP*(RDG)P*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA polymerase IV, ... | | Authors: | Nair, D.T, Kottur, J, Sharma, A. | | Deposit date: | 2014-04-13 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
6AA0
 
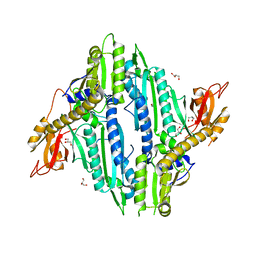 | | Crystal Structure of Toxoplasma gondii Prolyl tRNA Synthetase (TgPRS) in Apo Form | | Descriptor: | GLYCEROL, Prolyl-tRNA synthetase (ProRS) | | Authors: | Mishra, S, Kumari, S, Sharma, A, Yogavel, M. | | Deposit date: | 2018-07-16 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational heterogeneity in apo and drug-bound structures of Toxoplasma gondii prolyl-tRNA synthetase.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
4Q43
 
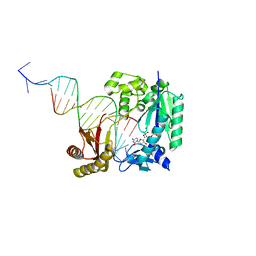 | | Polymerase-damaged DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*T*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*TP*CP*TP*(RDG)P*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Kottur, J, Sharma, A, Nair, D.T. | | Deposit date: | 2014-04-13 | | Release date: | 2015-05-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic
Structure, 23, 2015
|
|
3AFP
 
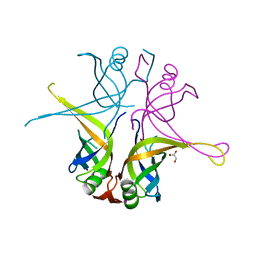 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KNP
 
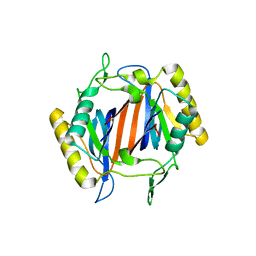 | |
3KOB
 
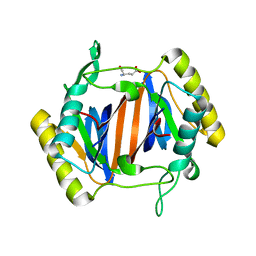 | |
3KO4
 
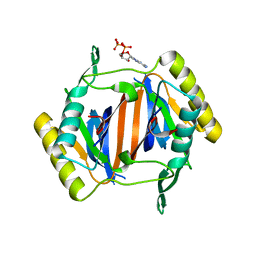 | |
3KOC
 
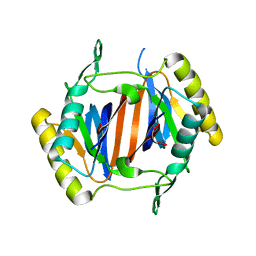 | |
3KNF
 
 | |
3KO7
 
 | |
3KO3
 
 | |
3KOD
 
 | |
3AFQ
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form II) | | Descriptor: | Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NSG
 
 | | Crystal Structure of OmpF, an Outer Membrane Protein from Salmonella typhi | | Descriptor: | CITRATE ANION, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Balasubramaniam, D, Arockiasamy, A, Sharma, A, Krishnaswamy, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Asymmetric pore occupancy in crystal structure of OmpF porin from Salmonella typhi
J.Struct.Biol., 178, 2012
|
|
5ZDL
 
 | | Crystal Structure Analysis of TtQRS in co-crystallised with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Glutamine--tRNA ligase | | Authors: | Mutharasappan, N, Jain, V, Sharma, A, Manickam, Y, Jeyaraman, J. | | Deposit date: | 2018-02-23 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of Glutaminyl-tRNA synthetase (TtGlnRS) from Thermus thermophilus HB8 and its complexes
Int. J. Biol. Macromol., 120, 2018
|
|
4IRD
 
 | | Structure of Polymerase-DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
3P5V
 
 | | Actinidin from Actinidia arguta planch (Sarusashi) | | Descriptor: | Actinidin, CADMIUM ION | | Authors: | Manickam, Y, Nirmal, N, Suzuki, A, Sugiyama, Y, Yamane, T, Devadasan, V, Sharma, A. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of actinidin and a comparison of cadmium and sulfur anomalous signals from actinidin crystals measured using in-house copper- and chromium-anode X-ray sources
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4IRC
 
 | | Polymerase-DNA complex | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), DNA (5'-D(*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
4IRK
 
 | | structure of Polymerase-DNA complex, dna | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*A*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*(DOC))-3'), DNA (5'-D(*TP*CP*TP*AP*GP*GP*GP*TP*CP*CP*TP*AP*GP*GP*AP*CP*CP*C)-3'), ... | | Authors: | Nair, D.T, Sharma, A. | | Deposit date: | 2013-01-14 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli.
Nucleic Acids Res., 41, 2013
|
|
3LMT
 
 | | Crystal structure of DTD from Plasmodium falciparum | | Descriptor: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | Authors: | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
