4ZPO
 
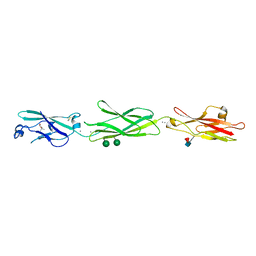 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Rubinstein, R, Honig, B, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
2OOX
 
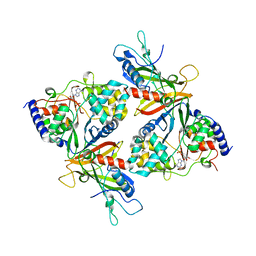 | |
2OOY
 
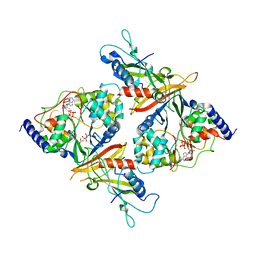 | |
4ZPL
 
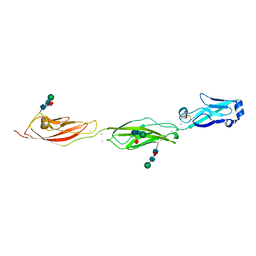 | | Crystal Structure of Protocadherin Beta 1 EC1-3 | | Descriptor: | CALCIUM ION, Protein Pcdhb1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Goodman, K.M, Bahna, F, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4ZPQ
 
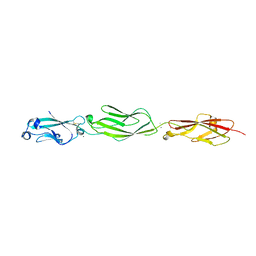 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
2QRD
 
 | | Crystal Structure of the Adenylate Sensor from AMP-activated Protein Kinase in complex with ADP and ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Protein C1556.08c, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
2QRE
 
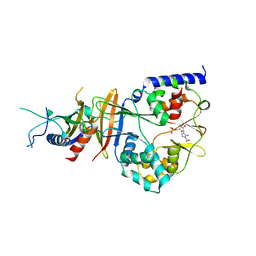 | | Crystal structure of the adenylate sensor from AMP-activated protein kinase in complex with 5-aminoimidazole-4-carboxamide 1-beta-D-ribofuranotide (ZMP) | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Protein C1556.08c, SNF1-like protein kinase ssp2, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
4ZPS
 
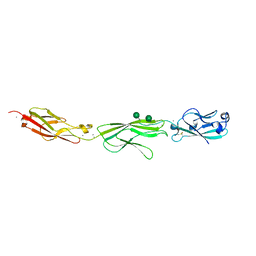 | | Crystal Structure of Protocadherin Gamma A8 EC1-3 | | Descriptor: | CALCIUM ION, MCG133388, isoform CRA_m, ... | | Authors: | Goodman, K.M, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4ZPN
 
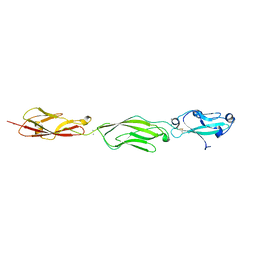 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 with extended N-terminus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Goodman, K.M, Wolcott, H.N, Bahna, F, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4ZPM
 
 | | Crystal Structure of Protocadherin Alpha C2 EC1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protein Pcdhac2, ... | | Authors: | Goodman, K.M, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
2QR1
 
 | | Crystal structure of the adenylate sensor from AMP-activated protein kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Protein C1556.08c, SNF1-like protein kinase ssp2, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-27 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
2NS9
 
 | | Crystal structure of protein APE2225 from Aeropyrum pernix K1, Pfam COXG | | Descriptor: | Hypothetical protein APE2225, PHOSPHATE ION | | Authors: | Jin, X, Bera, A, Wasserman, S, Smith, D, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-11-03 | | Release date: | 2006-11-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of hypothetical protein APE2225 from Aeropyrum pernix K1
To be Published
|
|
2QRC
 
 | | Crystal structure of the adenylate sensor from AMP-activated protein kinase in complex with ADP and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Protein C1556.08c, ... | | Authors: | Jin, X, Townley, R, Shapiro, L. | | Deposit date: | 2007-07-28 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insight into AMPK Regulation: ADP Comes into Play.
Structure, 15, 2007
|
|
4ZPP
 
 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
6VG1
 
 | | xenopus protocadherin 8.1 EC1-6 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Harrison, O.J, Brasch, B, Shapiro, L.S. | | Deposit date: | 2020-01-07 | | Release date: | 2020-03-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
2OCE
 
 | |
2V0G
 
 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A tRNA(leu) transcript with 5-FLUORO-1,3-DIHYDRO-1-HYDROXY-2,1- BENZOXABOROLE (AN2690) forming an adduct to the ribose of adenosine- 76 in the enzyme editing site. | | Descriptor: | AMINOACYL-TRNA SYNTHETASE, LEUCINE, MERCURY (II) ION, ... | | Authors: | Rock, F, Mao, W, Yaremchuk, A, Tukalo, M, Crepin, T, Zhou, H, Zhang, Y, Hernandez, V, Akama, T, Baker, S, Plattner, J, Shapiro, L, Martinis, S.A, Benkovic, S.J, Cusack, S, Alley, M.R.K. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An Antifungal Agent Inhibits an Aminoacyl-tRNA Synthetase by Trapping tRNA in the Editing Site.
Science, 316, 2007
|
|
2V0C
 
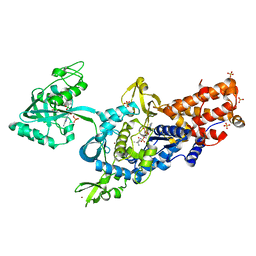 | | LEUCYL-TRNA SYNTHETASE FROM THERMUS THERMOPHILUS COMPLEXED WITH A SULPHAMOYL ANALOGUE OF LEUCYL-ADENYLATE In the synthetic site and an adduct of AMP with 5-Fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690) in the editing site | | Descriptor: | AMINOACYL-TRNA SYNTHETASE, LEUCINE, SULFATE ION, ... | | Authors: | Rock, F, Mao, W, Yaremchuk, A, Tukalo, M, Crepin, T, Zhou, H, Zhang, Y, Hernandez, V, Akama, T, Baker, S, Plattner, J, Shapiro, L, Martinis, S.A, Benkovic, S.J, Cusack, S, Alley, M.R.K. | | Deposit date: | 2007-05-14 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Antifungal Agent Inhibits an Aminoacyl-tRNA Synthetase by Trapping tRNA in the Editing Site.
Science, 316, 2007
|
|
5DZX
 
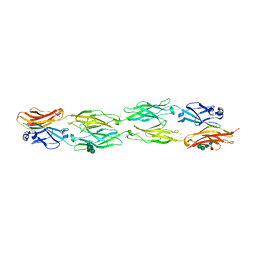 | | Protocadherin beta 6 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin beta 6, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.879 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
5DZW
 
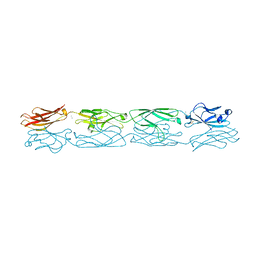 | | Protocadherin alpha 4 extracellular cadherin domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protocadherin alpha-4, ... | | Authors: | Goodman, K.M, Bahna, F, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
3LNE
 
 | |
5ERD
 
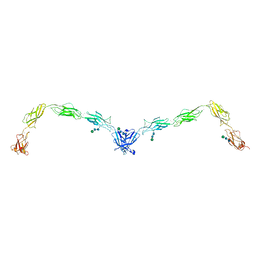 | | Crystal structure of human Desmoglein-2 ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brasch, J, Harrison, O.J, Shapiro, L. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5EQX
 
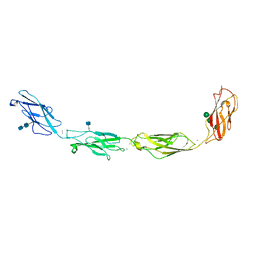 | | Crystal structure of human Desmoglein-3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3LNF
 
 | |
5EKE
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
