1SH5
 
 | |
1SAR
 
 | |
1T2H
 
 | |
3L1O
 
 | |
1ZGX
 
 | |
4J5G
 
 | |
4J5K
 
 | |
4JKQ
 
 | |
1BOX
 
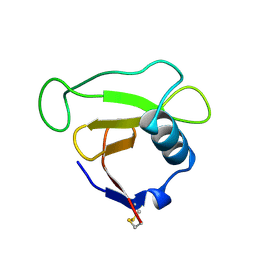 | | N39S MUTANT OF RNASE SA FROM STREPTOMYCES AUREOFACIENS | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA | | Authors: | Hebert, E.J, Giletto, A, Sevcik, J, Urbanikova, L, Wilson, K.S, Dauter, Z, Pace, C.N. | | Deposit date: | 1998-08-07 | | Release date: | 1999-12-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Contribution of a conserved asparagine to the conformational stability of ribonucleases Sa, Ba, and T1.
Biochemistry, 37, 1998
|
|
3D4A
 
 | |
3D5I
 
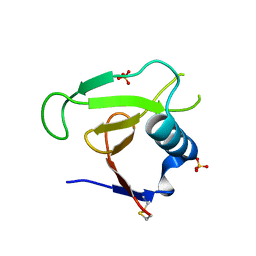 | |
3D5G
 
 | | Structure of ribonuclease Sa2 complexes with mononucleotides: new aspects of catalytic reaction and substrate recognition | | Descriptor: | Ribonuclease, SULFATE ION | | Authors: | Bauerova-Hlinkova, V, Dvorsky, R, Povazanec, F, Sevcik, J. | | Deposit date: | 2008-05-16 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of RNase Sa2 complexes with mononucleotides - new aspects of catalytic reaction and substrate recognition
Febs J., 276, 2009
|
|
1T2I
 
 | |
