5U25
 
 | |
5TVG
 
 | |
5U8O
 
 | |
5U7S
 
 | |
4Q14
 
 | |
5UXX
 
 | |
5VPV
 
 | |
5VWM
 
 | |
5IDV
 
 | |
5IDY
 
 | |
5J46
 
 | |
5IZ4
 
 | |
5WNN
 
 | |
5J6B
 
 | |
3D53
 
 | |
7JFZ
 
 | |
4O0K
 
 | |
8DOP
 
 | |
3DMO
 
 | |
8EGM
 
 | |
5KWV
 
 | |
3EK1
 
 | |
4DZ6
 
 | |
5B8I
 
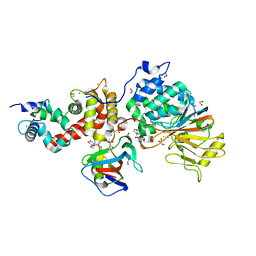 | | Crystal structure of Calcineurin A and Calcineurin B in complex with FKBP12 and FK506 from Coccidioides immitis RS | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID), Fox III, D, Dranow, D.M, Lorimer, D.D, Edwards, T.E. | | Deposit date: | 2015-05-03 | | Release date: | 2015-05-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Harnessing calcineurin-FK506-FKBP12 crystal structures from invasive fungal pathogens to develop antifungal agents.
Nat Commun, 10, 2019
|
|
3F0D
 
 | |
