6MAN
 
 | |
4W91
 
 | |
6MB1
 
 | | Crystal structure of N-myristoyl transferase (NMT) from Plasmodium vivax in complex with inhibitor IMP-1002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-{4-fluoro-2-[2-(1,3,5-trimethyl-1H-pyrazol-4-yl)ethoxy]phenyl}-1-methyl-1H-indazol-3-yl)-N,N-dimethylmethanamine, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-08-29 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Identification of Resistance Breaking Antimalarial N‐Myristoyltransferase Inhibitors.
Cell Chem Biol, 26, 2019
|
|
6E54
 
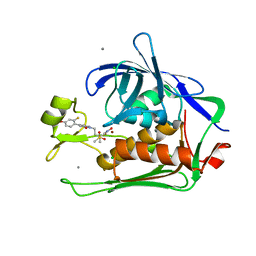 | |
6XDH
 
 | |
3V7O
 
 | |
7MYS
 
 | |
7MYQ
 
 | |
4POB
 
 | |
4PQ9
 
 | |
7N1L
 
 | |
7N7S
 
 | |
7SIQ
 
 | |
7SIR
 
 | |
7SH3
 
 | |
8UR4
 
 | |
8VQZ
 
 | |
8VQW
 
 | |
8W21
 
 | |
9AV6
 
 | |
9B0M
 
 | |
8W0J
 
 | |
8W0H
 
 | |
8UZN
 
 | |
9B21
 
 | | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form) | | Descriptor: | ADP-ribose pyrophosphatase, MAGNESIUM ION, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Seattle Structural Genomics Center for Infectious Disease, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2024-03-14 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ADP-ribose diphosphatase from Klebsiella pneumoniae (ADP Ribose bound, Orthorhombic P form)
To be published
|
|
