6QPZ
 
 | | Crystal structure of as isolated Y323E mutant of haem-Cu containing nitrite reductase from Ralstonia pickettii | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Antonyuk, S.V, Shenoy, R.T, Hedison, T.M, Eady, R.R, Hasnain, S.S, Scrutton, N.S. | | Deposit date: | 2019-02-16 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Unexpected Roles of a Tether Harboring a Tyrosine Gatekeeper Residue in Modular Nitrite Reductase Catalysis.
Acs Catalysis, 9, 2019
|
|
4HFJ
 
 | |
4GU5
 
 | | Structure of Full-length Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION | | Authors: | Zoltowski, B.D, Vaidya, A.T, Top, D, Widom, J, Young, M.W, Levy, C, Jones, A.R, Scrutton, N.S, Leys, D, Crane, B.R. | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Updated structure of Drosophila cryptochrome.
Nature, 495, 2013
|
|
4HFN
 
 | |
1O97
 
 | | Structure of electron transferring flavoprotein from Methylophilus methylotrophus, recognition loop removed by limited proteolysis | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
2XWZ
 
 | | STRUCTURE OF THE RECOMBINANT NATIVE NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | ACETATE ION, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
1O95
 
 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
2XX1
 
 | | STRUCTURE OF THE N90S MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
1O94
 
 | | Ternary complex between trimethylamine dehydrogenase and electron transferring flavoprotein | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ELECTRON TRANSFER FLAVOPROTEIN ALPHA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1O96
 
 | | Structure of electron transferring flavoprotein for Methylophilus methylotrophus. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ELECTRON TRANSFERRING FLAVOPROTEIN ALPHA-SUBUNIT, ELECTRON TRANSFERRING FLAVOPROTEIN BETA-SUBUNIT, ... | | Authors: | Leys, D, Basran, J, Talfournier, F, Sutcliffe, M.J, Scrutton, N.S. | | Deposit date: | 2002-12-11 | | Release date: | 2003-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Extensive Conformational Sampling in a Ternary Electron Transfer Complex.
Nat.Struct.Biol., 10, 2003
|
|
1PJ5
 
 | | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with acetate | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
1PJ7
 
 | | Structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folinic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, N,N-dimethylglycine oxidase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
2XX0
 
 | | STRUCTURE OF THE N90S-H254F MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
1PJ6
 
 | | Crystal structure of dimethylglycine oxidase of Arthrobacter globiformis in complex with folic acid | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FOLIC ACID, N,N-dimethylglycine oxidase, ... | | Authors: | Leys, D, Basran, J, Scrutton, N.S. | | Deposit date: | 2003-06-01 | | Release date: | 2003-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Channelling and formation of 'active' formaldehyde in dimethylglycine oxidase.
Embo J., 22, 2003
|
|
5L53
 
 | | Menthone neomenthol reductase from Mentha piperita in complex with NADP | | Descriptor: | (-)-menthone:(+)-neomenthol reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5L4S
 
 | | Isopiperitenone reductase from Mentha piperita in complex with NADP and beta-Cyclocitral | | Descriptor: | (-)-isopiperitenone reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, beta-cyclocitral | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-05-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5L51
 
 | |
5LDG
 
 | | Isopiperitenone reductase from Mentha piperita in complex with Isopiperitenone and NADP | | Descriptor: | (-)-Isopiperitenone, (-)-isopiperitenone reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-06-26 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
5LCX
 
 | | Isopiperitenone reductase from Mentha piperita in complex with NADP | | Descriptor: | (-)-isopiperitenone reductase, (4S)-2-METHYL-2,4-PENTANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Karuppiah, V, Toogood, H.S, Leys, D, Scrutton, N.S. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Pinpointing a Mechanistic Switch Between Ketoreduction and "Ene" Reduction in Short-Chain Dehydrogenases/Reductases.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3P62
 
 | |
3P80
 
 | |
4J2W
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot-Se) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
4J31
 
 | | Crystal Structure of kynurenine 3-monooxygenase (KMO-396Prot) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
3P7Y
 
 | |
4J36
 
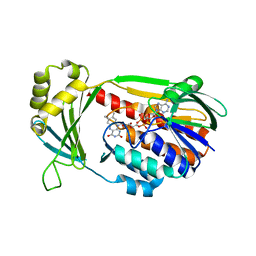 | | Cocrystal Structure of kynurenine 3-monooxygenase in complex with UPF 648 inhibitor(KMO-394UPF) | | Descriptor: | (1S,2S)-2-(3,4-dichlorobenzoyl)cyclopropanecarboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, Kynurenine 3-monooxygenase | | Authors: | Amaral, M, Levy, C, Heyes, D.J, Lafite, P, Outeiro, T.F, Giorgini, F, Leys, D, Scrutton, N.S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of kynurenine 3-monooxygenase inhibition.
Nature, 496, 2013
|
|
