2AGL
 
 | | Crystal structure of the phenylhydrazine adduct of aromatic amine dehydrogenase from Alcaligenes faecalis | | Descriptor: | 1-PHENYLHYDRAZINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AGY
 
 | | Crystal structure of the Schiff base intermediate in the reductive half-reaction of aromatic amine dehydrogenase (AADH) with tryptamine. Monoclinic form | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANIMINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
2AH0
 
 | | Crystal structure of the carbinolamine intermediate in the reductive half-reaction of aromatic amine dehydrogenase (AADH) with tryptamine. Monoclinic form | | Descriptor: | (1S)-1-AMINO-2-(1H-INDOL-3-YL)ETHANOL, 2-(1H-INDOL-3-YL)ETHANIMINE, Aromatic amine dehydrogenase | | Authors: | Masgrau, L, Roujeinikova, A, Johannissen, L.O, Hothi, P, Basran, J, Ranaghan, K.E, Mulholland, A.J, Sutcliffe, M.J, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-07-27 | | Release date: | 2006-04-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Atomic description of an enzyme reaction dominated by proton tunneling
Science, 312, 2006
|
|
3P8J
 
 | |
3P84
 
 | | Y351A mutant of pentaerythritol tetranitrate reductase containing a bound acetate molecule | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Toogood, H.S, Scrutton, N.S. | | Deposit date: | 2010-10-13 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Active site modifications in pentaerythritol tetranitrate reductase can lead to improved product enantiopurity, decreased by-product formation and altered stereochemical outcome in reactions with a,b-unsaturated nitroolefins
Catalysis Science and Technology, 2011
|
|
3P67
 
 | |
3P74
 
 | |
3P62
 
 | |
3P80
 
 | |
3P8I
 
 | | Y351F mutant of pentaerythritol tetranitrate reductase containing a bound acetate molecule | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Toogood, H.S, Scrutton, N.S. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Active site modifications in pentaerythritol tetranitrate reductase can lead to improved product enantiopurity, decreased by-product formation and altered stereochemical outcome in reactions with a,b-unsaturated nitroolefins
Catalysis Science and Technology, 2011
|
|
3P82
 
 | |
3P7Y
 
 | |
3P81
 
 | |
3GSI
 
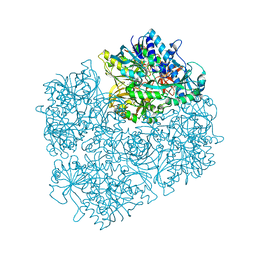 | | Crystal structure of D552A dimethylglycine oxidase mutant of Arthrobacter globiformis in complex with tetrahydrofolate | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tralau, T, Lafite, P, Levy, C, Combe, J.P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An internal reaction chamber in dimethylglycine oxidase provides efficient protection from exposure to toxic formaldehyde.
J.Biol.Chem., 284, 2009
|
|
3GX9
 
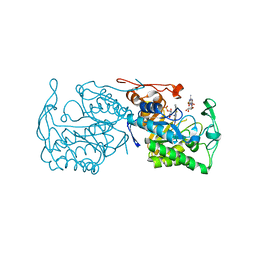 | | Structure of morphinone reductase N189A mutant in complex with tetrahydroNAD | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN MONONUCLEOTIDE, Morphinone reductase | | Authors: | Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-04-02 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Parallel Pathways and Free-Energy Landscapes for Enzymatic Hydride Transfer Probed by Hydrostatic Pressure
Chembiochem, 10, 2009
|
|
3IA5
 
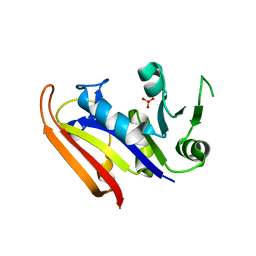 | | Moritella profunda dihydrofolate reductase (DHFR) | | Descriptor: | Dihydrofolate reductase, PHOSPHATE ION | | Authors: | Hay, S, Evans, R.M, Levy, C, Wang, X, Loveridge, E.J, Leys, D, Allemann, R.K, Scrutton, N.S. | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Are the Catalytic Properties of Enzymes from Piezophilic Organisms Pressure Adapted?
Chembiochem, 10, 2009
|
|
2XWZ
 
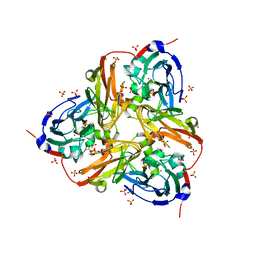 | | STRUCTURE OF THE RECOMBINANT NATIVE NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | ACETATE ION, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-06 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
2XX0
 
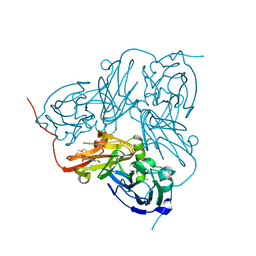 | | STRUCTURE OF THE N90S-H254F MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
3K8E
 
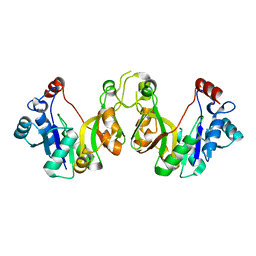 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase | | Descriptor: | 3-deoxy-manno-octulosonate cytidylyltransferase | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
2XX1
 
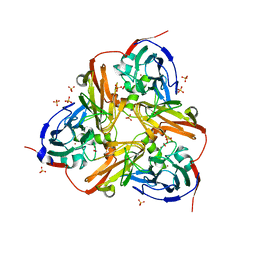 | | STRUCTURE OF THE N90S MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | Descriptor: | COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, NITRITE ION, ... | | Authors: | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | Deposit date: | 2010-11-07 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
3K8D
 
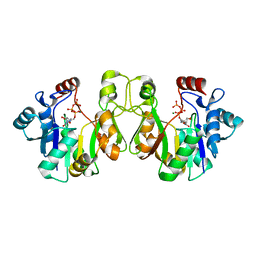 | | Crystal structure of E. coli lipopolysaccharide specific CMP-KDO synthetase in complex with CTP and 2-deoxy-Kdo | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, 3-deoxy-manno-octulosonate cytidylyltransferase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Heyes, D.J, Levy, C.W, Lafite, P, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based mechanism of CMP-2-keto-3-deoxymanno-octulonic acid synthetase: convergent evolution of a sugar-activating enzyme with DNA/RNA polymerases
J.Biol.Chem., 284, 2009
|
|
3KP0
 
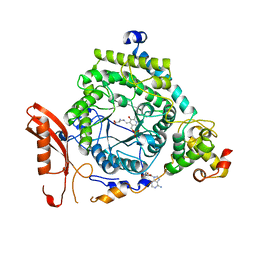 | | Crystal Structure of ORNITHINE 4,5 AMINOMUTASE in complex with 2,4-diaminobutyrate (DAB) (Aerobic) | | Descriptor: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KFT
 
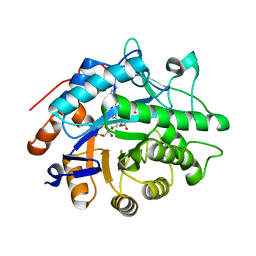 | | Crystal structure of Pentaerythritol Tetranitrate Reductase complex with 1,4,5,6-tetrahydro NADH | | Descriptor: | FLAVIN MONONUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pentaerythritol tetranitrate reductase | | Authors: | Pudney, C.R, Levy, C.W, Leys, D, Scrutton, N.S. | | Deposit date: | 2009-10-28 | | Release date: | 2010-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evidence to support the hypothesis that promoting vibrations enhance the rate of an enzyme catalyzed H-tunneling reaction.
J.Am.Chem.Soc., 131, 2009
|
|
3KOY
 
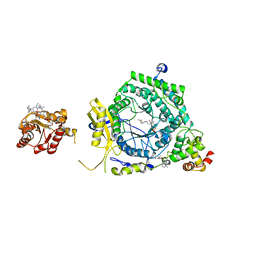 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Aerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
