4KOA
 
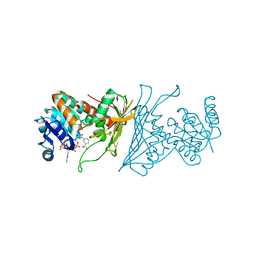 | | Crystal Structure Analysis of 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti | | Descriptor: | 1,5-anhydro-D-fructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schu, M, Faust, A, Stosik, B, Kohring, G.-W, Giffhorn, F, Scheidig, A.J. | | Deposit date: | 2013-05-11 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The structure of substrate-free 1,5-anhydro-D-fructose reductase from Sinorhizobium meliloti 1021 reveals an open enzyme conformation.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6DXO
 
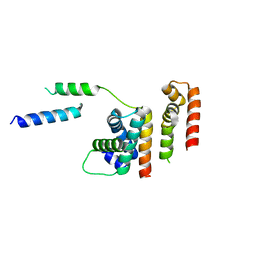 | | 1.8 A structure of RsbN-BldN complex. | | Descriptor: | BldN, RNA polymerase ECF-subfamily sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-06-29 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the RsbN-sigma BldN complex from Streptomyces venezuelae defines a new structural class of anti-sigma factor.
Nucleic Acids Res., 46, 2018
|
|
3TPE
 
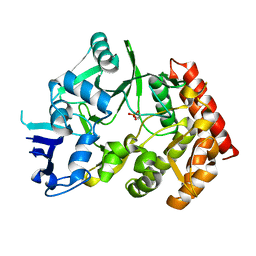 | | The phipa p3121 structure | | Descriptor: | Serine/threonine-protein kinase HipA | | Authors: | Schumacher, M.A, Link, T, Brennan, R.G. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
7RMW
 
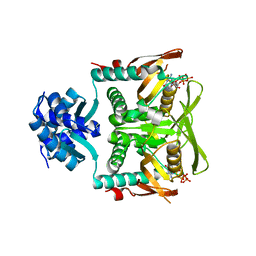 | | Crystal structure of B. subtilis PurR bound to ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, Pur operon repressor | | Authors: | Schumacher, M.A. | | Deposit date: | 2021-07-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The nucleotide messenger (p)ppGpp is an anti-inducer of the purine synthesis transcription regulator PurR in Bacillus.
Nucleic Acids Res., 50, 2022
|
|
4W4G
 
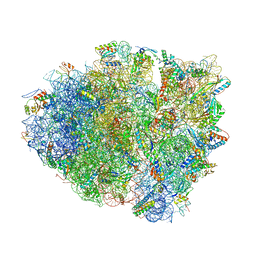 | | Postcleavage state of 70S bound to HigB toxin and AAA (lysine) codon | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Schureck, M.A, Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2014-08-14 | | Release date: | 2015-10-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Defining the mRNA recognition signature of a bacterial toxin protein.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6E4N
 
 | |
6E4P
 
 | | Structure of the T. brucei RRM domain in complex with RNA | | Descriptor: | RNA (5'-R(P*UP*UP*UP*U)-3'), RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
5I44
 
 | | Structure of RacA-DNA complex; P21 form | | Descriptor: | Chromosome-anchoring protein RacA, DNA (5'-D(*TP*GP*AP*CP*GP*CP*CP*GP*GP*CP*GP*TP*CP*A)-3') | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-02-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.621 Å) | | Cite: | Molecular insights into DNA binding and anchoring by the Bacillus subtilis sporulation kinetochore-like RacA protein.
Nucleic Acids Res., 44, 2016
|
|
5I98
 
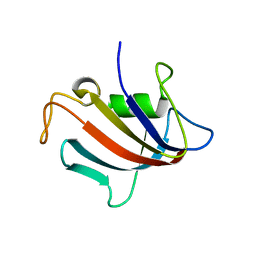 | |
6E4O
 
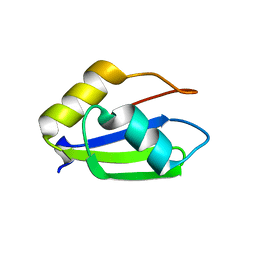 | | Structure of apo T. brucei RRM: P4(1)2(1)2 form | | Descriptor: | RNA-binding protein, putative | | Authors: | Schumacher, M.A. | | Deposit date: | 2018-07-18 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The RRM of the kRNA-editing protein TbRGG2 uses multiple surfaces to bind and remodel RNA.
Nucleic Acids Res., 47, 2019
|
|
5HAW
 
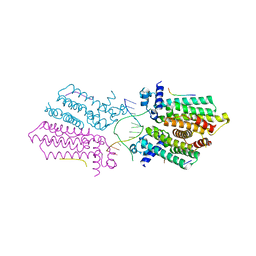 | |
2PUD
 
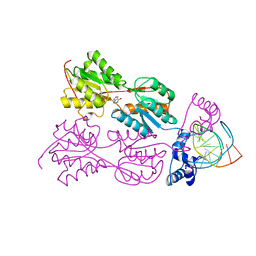 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
2PUC
 
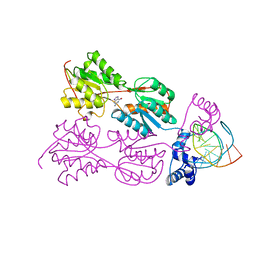 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
1G4Y
 
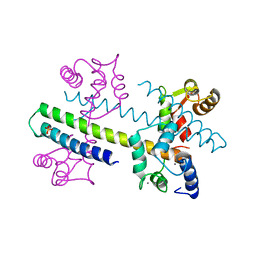 | | 1.60 A CRYSTAL STRUCTURE OF THE GATING DOMAIN FROM SMALL CONDUCTANCE POTASSIUM CHANNEL COMPLEXED WITH CALCIUM-CALMODULIN | | Descriptor: | CALCIUM ION, CALCIUM-ACTIVATED POTASSIUM CHANNEL RSK2, CALMODULIN, ... | | Authors: | Schumacher, M.A, Rivard, A, Bachinger, H.P, Adelman, J.P. | | Deposit date: | 2001-01-07 | | Release date: | 2001-05-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin.
Nature, 410, 2001
|
|
5HT1
 
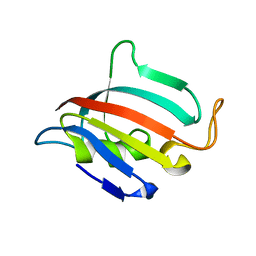 | | Structure of apo C. glabrata FKBP12 | | Descriptor: | FK506-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-01-26 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structures of Pathogenic Fungal FKBP12s Reveal Possible Self-Catalysis Function.
Mbio, 7, 2016
|
|
5HUA
 
 | | Structure of C. glabrata FKBP12-FK506 complex | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506-binding protein 1 | | Authors: | Schumacher, M.A. | | Deposit date: | 2016-01-27 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of Pathogenic Fungal FKBP12s Reveal Possible Self-Catalysis Function.
Mbio, 7, 2016
|
|
5HTG
 
 | |
2Q2K
 
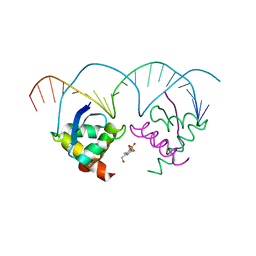 | | Structure of nucleic-acid binding protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DNA (5'-D(*AP*GP*TP*AP*TP*AP*(5IU)P*AP*CP*(5IU)P*AP*GP*TP*AP*TP*AP*TP*AP*CP*T)-3'), Hypothetical protein | | Authors: | Schumacher, M.A, Glover, T, Firth, N. | | Deposit date: | 2007-05-28 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Segrosome structure revealed by a complex of ParR with centromere DNA.
Nature, 450, 2007
|
|
4RS8
 
 | |
8SVD
 
 | |
6PFJ
 
 | | Structure of S. venezuelae RsiG-WhiG-(ci-di-GMP) complex, P64 crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-21 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6PFV
 
 | | Structure of S. venezuelae RisG-WhiG-c-di-GMP complex: orthorhombic crystal form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), AmfC protein, RNA polymerase sigma factor | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-06-22 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | c-di-GMP Arms an Anti-sigma to Control Progression of Multicellular Differentiation in Streptomyces.
Mol.Cell, 77, 2020
|
|
6NOY
 
 | | Structure of Cyanothece McdB | | Descriptor: | Maintenance of carboxysome positioning B protein, Mcsb | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-24 | | Last modified: | 2019-06-26 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of maintenance of carboxysome distribution Walker-box McdA and McdB adaptor homologs.
Nucleic Acids Res., 47, 2019
|
|
6NL1
 
 | | Structure of T. brucei MERS1 protein in its apo form | | Descriptor: | Mitochondrial edited mRNA stability factor 1, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
6NOO
 
 | | Structure of Cyanothece McdA-AMPPNP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Maintenance of carboxysome positioning A protein, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-01-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of maintenance of carboxysome distribution Walker-box McdA and McdB adaptor homologs.
Nucleic Acids Res., 47, 2019
|
|
