3ZQ1
 
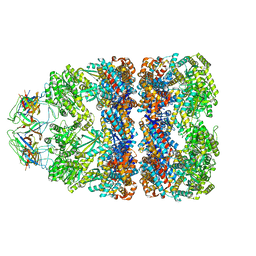 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15.9 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
4A13
 
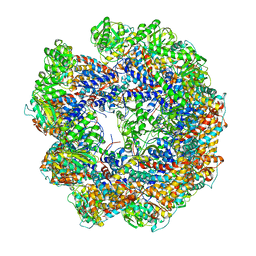 | | model refined against symmetry-free cryo-EM map of TRiC-ADP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0W
 
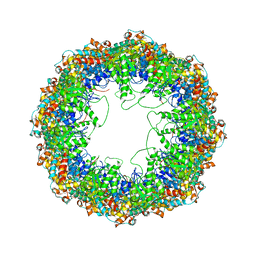 | | model built against symmetry-free cryo-EM map of TRiC-ADP-AlFx | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.9 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
4A0V
 
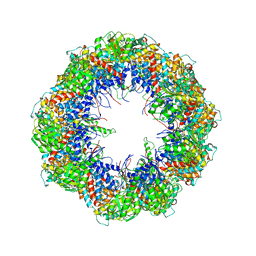 | | model refined against the Symmetry-free cryo-EM map of TRiC-AMP-PNP | | Descriptor: | T-COMPLEX PROTEIN 1 SUBUNIT BETA | | Authors: | Cong, Y, Schroder, G.F, Meyer, A.S, Jakana, J, Ma, B, Dougherty, M.T, Schmid, M.F, Reissmann, S, Levitt, M, Ludtke, S.L, Frydman, J, Chiu, W. | | Deposit date: | 2011-09-13 | | Release date: | 2012-02-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Symmetry-Free Cryo-Em Structures of the Chaperonin Tric Along its ATPase-Driven Conformational Cycle.
Embo J., 31, 2012
|
|
3J63
 
 | | Unified assembly mechanism of ASC-dependent inflammasomes | | Descriptor: | Apoptosis-associated speck-like protein containing a CARD | | Authors: | Lu, A, Magupalli, V.G, Ruan, J, Yin, Q, Atianand, M.K, Vos, M, Schroder, G.F, Fitzgerald, K.A, Wu, H, Egelman, E.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes.
Cell(Cambridge,Mass.), 156, 2014
|
|
3J8K
 
 | | Tilted state of actin, T2 | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Galkin, V.E, Orlova, A, Vos, M.R, Schroder, G.F, Egelman, E.H. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Near-atomic resolution for one state of f-actin.
Structure, 23, 2015
|
|
3J8J
 
 | | Tilted state of actin, T1 | | Descriptor: | Actin, alpha skeletal muscle | | Authors: | Galkin, V.E, Orlova, A, Vos, M.R, Schroder, G.F, Egelman, E.H. | | Deposit date: | 2014-11-07 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Near-atomic resolution for one state of f-actin.
Structure, 23, 2015
|
|
3TX8
 
 | | Crystal structure of a succinyl-diaminopimelate desuccinylase (ArgE) from Corynebacterium glutamicum ATCC 13032 at 2.97 A resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Succinyl-diaminopimelate desuccinylase | | Authors: | Joint Center for Structural Genomics (JCSG), Brunger, A.T, Terwilliger, T.C, Read, R.J, Adams, P.D, Levitt, M, Schroder, G.F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.972 Å) | | Cite: | Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing: the structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6Y1A
 
 | | Amyloid fibril structure of islet amyloid polypeptide | | Descriptor: | AMINO GROUP, Islet amyloid polypeptide | | Authors: | Roeder, C, Kupreichyk, T, Gremer, L, Schaefer, L.U, Pothula, K.R, Ravelli, R.B.G, Willbold, D, Hoyer, W, Schroder, G.F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-beta fibrils.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6YEG
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - Final EM Refinement | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-03-24 | | Release date: | 2020-10-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
6YQ5
 
 | | Hybrid structure of the SPP1 tail tube by solid-state NMR and cryo EM - NMR Ensemble | | Descriptor: | Tail tube protein gp17.1* | | Authors: | Zinke, M, Sachowsky, K.A.A, Zinn-Justin, S, Ravelli, R, Schroder, G.F, Habeck, M, Lange, A. | | Deposit date: | 2020-04-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å), SOLID-STATE NMR | | Cite: | Architecture of the flexible tail tube of bacteriophage SPP1.
Nat Commun, 11, 2020
|
|
