5V7Y
 
 | |
8UF8
 
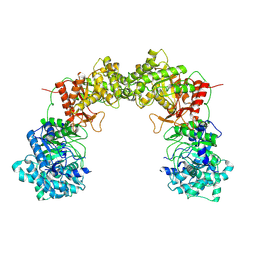 | | Cryo-EM structure of alpha-Klotho | | Descriptor: | Klotho | | Authors: | Schnicker, N.J, Xu, Z, Mohammad, A, Gakhar, L, Huang, C.L. | | Deposit date: | 2023-10-03 | | Release date: | 2024-10-16 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Conformational landscape of soluble alpha-klotho revealed by cryogenic electron microscopy.
Sci Rep, 15, 2025
|
|
8TKQ
 
 | |
8TOH
 
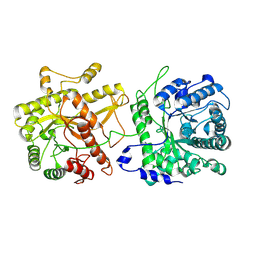 | | Cryo-EM structure of monomeric alpha-Klotho | | Descriptor: | Klotho | | Authors: | Schnicker, N.J, Xu, Z, Mohammad, A, Gakhar, L, Huang, C.L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-14 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Conformational landscape of soluble alpha-klotho revealed by cryogenic electron microscopy.
Sci Rep, 15, 2025
|
|
5HV0
 
 | |
5HV4
 
 | |
5IAX
 
 | |
5IAT
 
 | |
5IAV
 
 | |
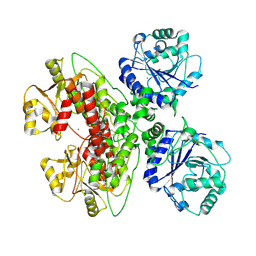
7UI7
 
 | |
5TG0
 
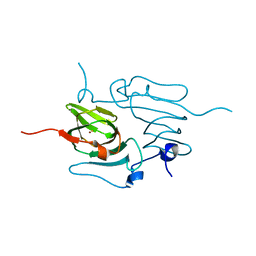 | |
5TFZ
 
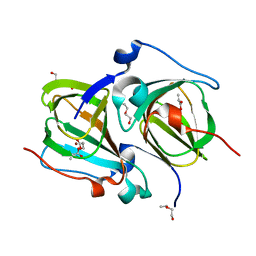 | |
7JXV
 
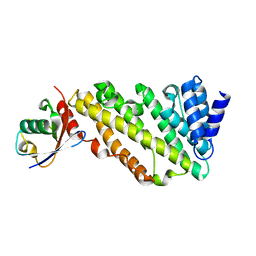 | | ANTH domain of CALM (clathrin-assembly lymphoid myeloid leukemia protein) bound to ubiquitin | | Descriptor: | Phosphatidylinositol-binding clathrin assembly protein, Ubiquitin | | Authors: | Pashkova, N, Gakhar, L, Schnicker, N.J, Piper, R.C. | | Deposit date: | 2020-08-28 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | ANTH domains within CALM, HIP1R, and Sla2 recognize ubiquitin internalization signals.
Elife, 10, 2021
|
|
9DVQ
 
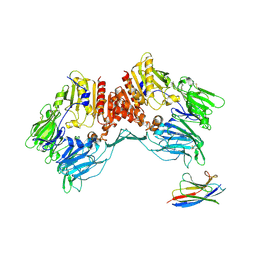 | | Cryo-EM structure of Human Fibroblast Activation Protein alpha dimer with one SUMO-I3 VHHs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiplasmin-cleaving enzyme FAP, soluble form, ... | | Authors: | Xu, Z, Schnicker, N.J, Wadas, T.J. | | Deposit date: | 2024-10-08 | | Release date: | 2025-02-26 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-electron microscopy reveals a single domain antibody with a unique binding epitope on fibroblast activation protein alpha.
Rsc Chem Biol, 6, 2025
|
|
9DVR
 
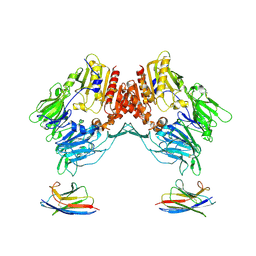 | | Cryo-EM structure of Human Fibroblast Activation Protein alpha dimer with two SUMO-I3 VHHs bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antiplasmin-cleaving enzyme FAP, soluble form, ... | | Authors: | Xu, Z, Schnicker, N.J, Wadas, T.J. | | Deposit date: | 2024-10-08 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-electron microscopy reveals a single domain antibody with a unique binding epitope on fibroblast activation protein alpha.
Rsc Chem Biol, 6, 2025
|
|
8U0I
 
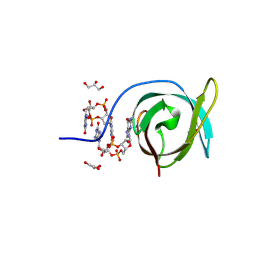 | | Crystal structure of PA0012 complexed with cyclic-di-GMP from Pseudomonas aeruginosa | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), GLYCEROL, PilZ domain-containing protein | | Authors: | Hammons, N.A, Schnicker, N.J, Fuentes, E.J. | | Deposit date: | 2023-08-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Crystal structure of PA0012 complexed with cyclic-di-GMP from Pseudomonas aeruginosa
To Be Published
|
|
9C8S
 
 | |
9C8R
 
 | |
7UI6
 
 | |
