9GIL
 
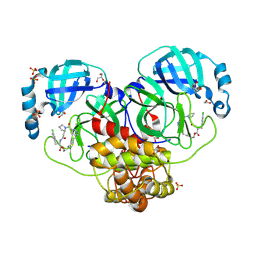 | | Crystal structure of SARS-CoV-2 Mpro with compound 12 | | Descriptor: | (7~{R},11~{R},19~{E})-11-[(4-chlorophenyl)methyl]-13-oxa-3,10,23-triazatricyclo[19.3.1.0^{3,7}]pentacosa-1(24),19,21(25),22-tetraene-2,9,12-trione, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Schmitt, A, Preuss, F, Prasad, A, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 2024
|
|
8QQG
 
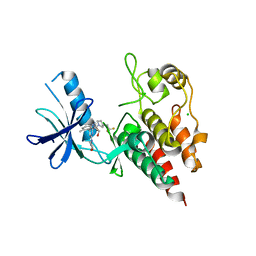 | | Structure of BRAF in Complex With Exarafenib (KIN-2787). | | Descriptor: | (3~{S})-~{N}-[4-methyl-3-[2-morpholin-4-yl-6-[[(2~{R})-1-oxidanylpropan-2-yl]amino]pyridin-4-yl]phenyl]-3-[2,2,2-tris(fluoranyl)ethyl]pyrrolidine-1-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | Authors: | Schmitt, A, Costanzi, E, Kania, R, Chen, Y.K. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.979 Å) | | Cite: | Structure of BRAF in Complex With Exarafenib (KIN-2787).
To Be Published
|
|
6EVU
 
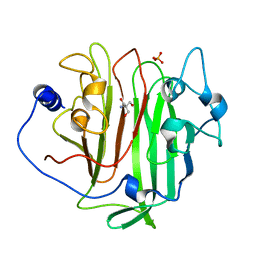 | | Adhesin domain of PrgB from Enterococcus faecalis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PHOSPHATE ION, PrgB | | Authors: | Schmitt, A, Berntsson, R.P.A. | | Deposit date: | 2017-11-02 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction.
Mol. Microbiol., 109, 2018
|
|
6GED
 
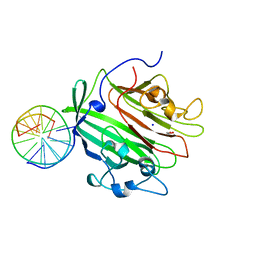 | | Adhesin domain of PrgB from Enterococcus faecalis bound to DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(P*CP*GP*GP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(P*GP*GP*GP*CP*GP*GP*CP*CP*CP*G)-3'), ... | | Authors: | Schmitt, A, Berntsson, R.P.A. | | Deposit date: | 2018-04-26 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | PrgB promotes aggregation, biofilm formation, and conjugation through DNA binding and compaction.
Mol. Microbiol., 109, 2018
|
|
6FA9
 
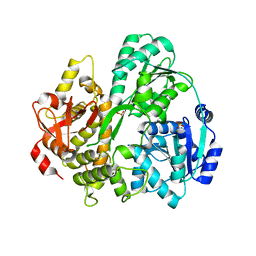 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 | | Descriptor: | Putative mRNA splicing factor, SULFATE ION | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAC
 
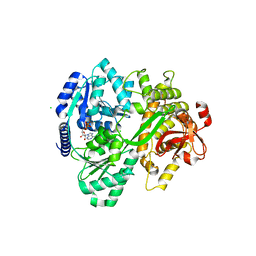 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6FAA
 
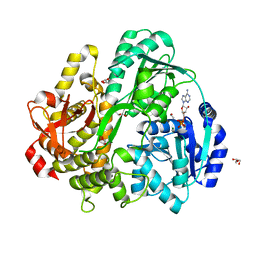 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ETHANOL, GLYCEROL, ... | | Authors: | Schmitt, A, Hamann, F, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6Z9L
 
 | | Enterococcal PrgA | | Descriptor: | Poly-alanine peptide, PrgA, SULFATE ION | | Authors: | Berntsson, R.P.A, Schmitt, A. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.063 Å) | | Cite: | Enterococcal PrgA Extends Far Outside the Cell and Provides Surface Exclusion to Protect against Unwanted Conjugation.
J.Mol.Biol., 432, 2020
|
|
7PTV
 
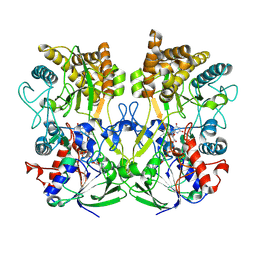 | | Structure of the Mimivirus genomic fibre asymmetric unit | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2021-09-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
8ORH
 
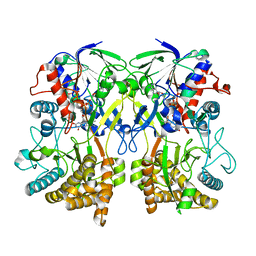 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
8ORS
 
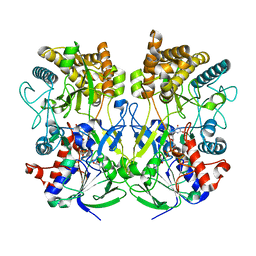 | | Knockout of GMC-oxidoreductase genes reveals that functional redundancy preserves mimivirus essential functions | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Alempic, J.M, Bisio, H, Villalta, A, Santini, S, Lartigue, A, Schmitt, A, Bugnot, C, Notaro, A, Belmudes, L, Adrait, A, Poirot, O, Ptchelkine, D, De Castro, C, Coute, Y, Abergel, C. | | Deposit date: | 2023-04-17 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional redundancy revealed by the deletion of the mimivirus GMC-oxidoreductase genes.
Microlife, 5, 2024
|
|
7YX3
 
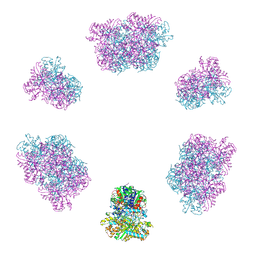 | | Structure of the Mimivirus genomic fibre in its compact 6-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative GMC-type oxidoreductase | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30-nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX4
 
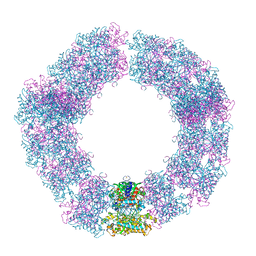 | | Structure of the Mimivirus genomic fibre in its compact 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
7YX5
 
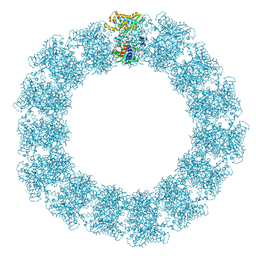 | | Structure of the Mimivirus genomic fibre in its relaxed 5-start helix form | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative glucose-methanol-choline oxidoreductase protein | | Authors: | Villalta, A, Schmitt, A, Estrozi, L.F, Quemin, E.R.J, Alempic, J.M, Lartigue, A, Prazak, V, Belmudes, L, Vasishtan, D, Colmant, A.M.G, Honore, F.A, Coute, Y, Grunewald, K, Abergel, C. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The giant mimivirus 1.2 Mb genome is elegantly organized into a 30 nm diameter helical protein shield.
Elife, 11, 2022
|
|
9GIJ
 
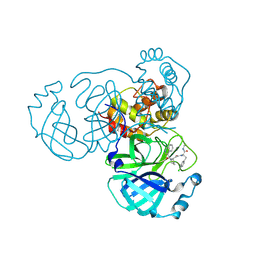 | | Crystal structure of SARS-CoV-2 Mpro with compound 5 | | Descriptor: | (2~{R})-3-(4-chlorophenyl)-2-[2-[(2~{R})-1-isoquinolin-4-ylcarbonylpyrrolidin-2-yl]ethanoyl-methyl-amino]-~{N}-methyl-propanamide, 3C-like proteinase nsp5 | | Authors: | Prasad, A, Schmitt, A, Preuss, F, Maskos, K, Wang, X, Gotchev, D, Konz Makino, D.L. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Rational Design of Macrocyclic Noncovalent Inhibitors of SARS-CoV-2 M pro from a DNA-Encoded Chemical Library Screening Hit That Demonstrate Potent Inhibition against Pan-Coronavirus Homologues and Nirmatrelvir-Resistant Variants.
J.Med.Chem., 2024
|
|
6Z9K
 
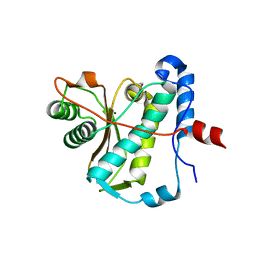 | | CAP domain of Enterococcal PrgA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, PrgA | | Authors: | Berntsson, R.P.A, Schmitt, A. | | Deposit date: | 2020-06-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enterococcal PrgA Extends Far Outside the Cell and Provides Surface Exclusion to Protect against Unwanted Conjugation.
J.Mol.Biol., 432, 2020
|
|
8BEG
 
 | | Structure of Ig-like domains from PrgB | | Descriptor: | MAGNESIUM ION, PrgB | | Authors: | Jarva, M, Schmitt, A, Berntsson, R.P.-A. | | Deposit date: | 2022-10-21 | | Release date: | 2022-11-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural foundation for the role of enterococcal PrgB in conjugation, biofilm formation, and virulence.
Elife, 12, 2023
|
|
6FA5
 
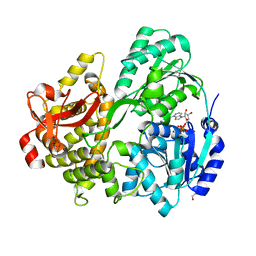 | | CRYSTAL STRUCTURE OF THE DEAH-BOX HELICASE PRP2 IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Hamann, F, Schmitt, A, Neumann, P, Ficner, R. | | Deposit date: | 2017-12-15 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Crystal structure of the spliceosomal DEAH-box ATPase Prp2.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6RMC
 
 | | Crystal structure of the DEAH-box ATPase Prp2 in complex with Spp2 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative mRNA splicing factor, ... | | Authors: | Hamann, F, Neumann, P, Schmitt, A, Ficner, R. | | Deposit date: | 2019-05-06 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of the intrinsically disordered splicing factor Spp2 and its binding to the DEAH-box ATPase Prp2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
