1PHO
 
 | |
3PK2
 
 | | Artificial Transfer Hydrogenases for the Enantioselective Reduction of Cyclic Imines | | Descriptor: | IRIDIUM (III) ION, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Schirmer, T, Heinisch, T. | | Deposit date: | 2010-11-11 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Artificial transfer hydrogenases for the enantioselective reduction of cyclic imines.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
4OKA
 
 | | Structural-, Kinetic- and Docking Studies of Artificial Imine Reductases Based on the Biotin-Streptavidin Technology: An Induced Lock-and-Key Hypothesis | | Descriptor: | IRIDIUM ION, Streptavidin, [N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-(2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)pentanamidato]iridium(III) | | Authors: | Schirmer, T, Heinisch, T. | | Deposit date: | 2014-01-22 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Structural, Kinetic, and Docking Studies of Artificial Imine Reductases Based on Biotin-Streptavidin Technology: An Induced Lock-and-Key Hypothesis
J.Am.Chem.Soc., 136, 2014
|
|
2BK0
 
 | |
1MPF
 
 | |
1MAL
 
 | |
2WPU
 
 | | Chaperoned ruthenium metallodrugs that recognize telomeric DNA | | Descriptor: | (3AS,4S,6AR)-4-(5-((3R,4R)-3,4-DIAMINOPYRROLIDIN-1-YL)-5-OXOPENTYL)TETRAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-2(3H)-ONE-P-CYMENE-CHLORO-RUTHENIUM(III), GLYCEROL, STREPTAVIDIN, ... | | Authors: | Heinisch, T, Schirmer, T, Zimbron, J.M, Sardo, A, Wohlschlager, T, Gradinaru, J, Creus, M, Ward, T.R. | | Deposit date: | 2009-08-10 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Chemo-Genetic Optimization of DNA Recognition by Metallodrugs Using a Presenter-Protein Strategy.
Chemistry, 16, 2010
|
|
8BVB
 
 | |
3T9O
 
 | | Regulatory CZB domain of DgcZ | | Descriptor: | Diguanylate cyclase DgcZ, ZINC ION | | Authors: | Zaehringer, F, Schirmer, T. | | Deposit date: | 2011-08-03 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and signaling mechanism of a zinc-sensory diguanylate cyclase.
Structure, 21, 2013
|
|
2JK8
 
 | |
7B0E
 
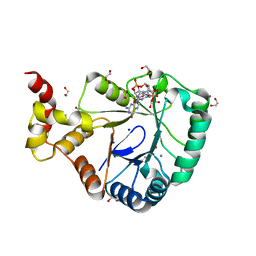 | | Crystal structure of SmbA loop deletion mutant | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, ... | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structure of loop deletion mutant SmbA bound to a monomeric c-di-GMP
To Be Published
|
|
2WQL
 
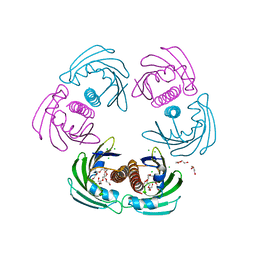 | | CRYSTAL STRUCTURE OF THE MAJOR CARROT ALLERGEN DAU C 1 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAJOR ALLERGEN DAU C 1, ... | | Authors: | Markovic-Housley, Z, Basle, A, Padavattan, S, Hoffmann-Sommergruber, K, Schirmer, T. | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the Major Carrot Allergen Dau C 1.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
1HXT
 
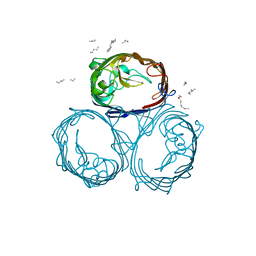 | | OMPF PORIN MUTANT NQAAA | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, OUTER MEMBRANE PROTEIN F | | Authors: | Phale, P.S, Philippsen, A, Widmer, C, Phale, V.P, Rosenbusch, J.P, Schirmer, T. | | Deposit date: | 2001-01-17 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation.
Biochemistry, 40, 2001
|
|
1HXX
 
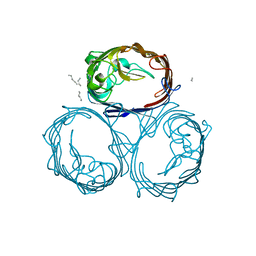 | | OMPF PORIN MUTANT Y106F | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, OUTER MEMBRANE PROTEIN F | | Authors: | Phale, P.S, Philippsen, A, Widmer, C, Phale, V.P, Rosenbusch, J.P, Schirmer, T. | | Deposit date: | 2001-01-17 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation.
Biochemistry, 40, 2001
|
|
1HXU
 
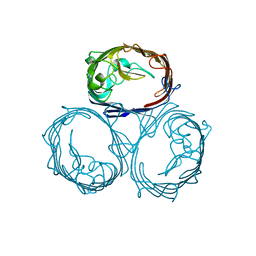 | | OMPF PORIN MUTANT KK | | Descriptor: | OUTER MEMBRANE PROTEIN F | | Authors: | Phale, P.S, Philippsen, A, Widmer, C, Phale, V.P, Rosenbusch, J.P, Schirmer, T. | | Deposit date: | 2001-01-17 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation.
Biochemistry, 40, 2001
|
|
7PK5
 
 | |
6ZXM
 
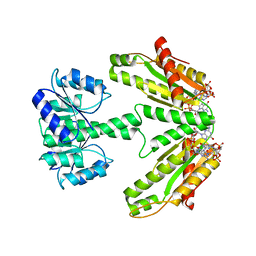 | | Diguanylate cyclase DgcR in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Putative GGDEF/response regulator receiver domain protein | | Authors: | Teixeira, R.D, Schirmer, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Activation mechanism of a small prototypic Rec-GGDEF diguanylate cyclase.
Nat Commun, 12, 2021
|
|
6ZXC
 
 | |
6ZXB
 
 | |
5NH2
 
 | |
1AF6
 
 | | MALTOPORIN SUCROSE COMPLEX | | Descriptor: | MAGNESIUM ION, MALTOPORIN, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Dutzler, R, Schirmer, T. | | Deposit date: | 1997-03-21 | | Release date: | 1998-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin.
J.Mol.Biol., 272, 1997
|
|
6TNE
 
 | |
5IDJ
 
 | |
4XJY
 
 | | Periplasmic repressor protein YfiR | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, L(+)-TARTARIC ACID, YfiR | | Authors: | Kauer, S, Schirmer, T. | | Deposit date: | 2015-01-09 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Periplasmic repressor protein YfiR at 1.8 Angstroms resolution
To Be Published
|
|
6GS8
 
 | | Crystal structure of SmbA in complex with c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MAGNESIUM ION, Uncharacterized protein | | Authors: | Dubey, B.N, Schirmer, T. | | Deposit date: | 2018-06-13 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Reciprocal growth control by competitive binding of nucleotide second messengers to a metabolic switch in Caulobacter crescentus
Nat Microbiol, 2021
|
|
