6I71
 
 | |
4S3P
 
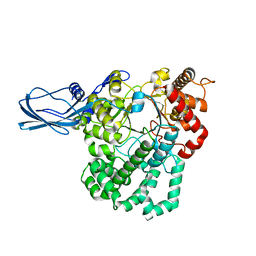 | |
4S3Q
 
 | | Amylomaltase MalQ from Escherichia coli in complex with maltose | | 分子名称: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase, IODIDE ION, ... | | 著者 | Weiss, S.C, Schiefner, A. | | 登録日 | 2015-03-26 | | 公開日 | 2015-07-08 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for the Interconversion of Maltodextrins by MalQ, the Amylomaltase of Escherichia coli.
J.Biol.Chem., 290, 2015
|
|
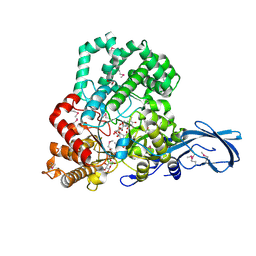
4S3R
 
 | |
3CJJ
 
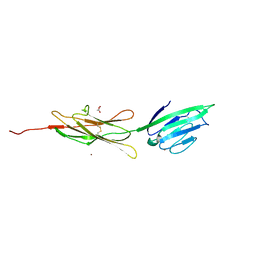 | | Crystal structure of human rage ligand-binding domain | | 分子名称: | ACETATE ION, Advanced glycosylation end product-specific receptor, ZINC ION | | 著者 | Koch, M, Dattilo, B.M, Schiefner, A, Diez, J, Chazin, W.J, Fritz, G. | | 登録日 | 2008-03-13 | | 公開日 | 2009-03-24 | | 最終更新日 | 2011-12-28 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural basis for ligand recognition and activation of RAGE.
Structure, 18, 2010
|
|
1URS
 
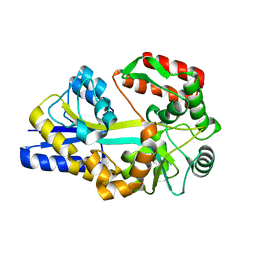 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | 分子名称: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | 登録日 | 2003-11-04 | | 公開日 | 2003-12-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins
J.Mol.Biol., 335, 2004
|
|
1URD
 
 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius provide insight into acid stability of proteins | | 分子名称: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | 登録日 | 2003-10-29 | | 公開日 | 2003-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.53 Å) | | 主引用文献 | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1URG
 
 | | X-ray structures from the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | 分子名称: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | 著者 | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | 登録日 | 2003-10-29 | | 公開日 | 2003-12-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
4I5E
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase in complex with NADP+ | | 分子名称: | Alclohol dehydrogenase/short-chain dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | 登録日 | 2012-11-28 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5D
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase in its apo form | | 分子名称: | Alclohol dehydrogenase/short-chain dehydrogenase, SULFATE ION | | 著者 | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | 登録日 | 2012-11-28 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5G
 
 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S, A86N, S88A | | 分子名称: | Alclohol dehydrogenase/short-chain dehydrogenase | | 著者 | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | 登録日 | 2012-11-28 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4I5F
 
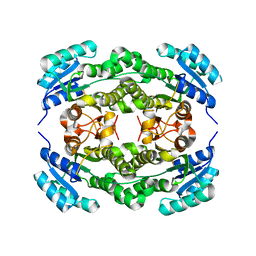 | | Crystal structure of Ralstonia sp. alcohol dehydrogenase mutant N15G, G37D, R38V, R39S | | 分子名称: | Alclohol dehydrogenase/short-chain dehydrogenase | | 著者 | Jarasch, A, Lerchner, A, Meining, W, Schiefner, A, Skerra, A. | | 登録日 | 2012-11-28 | | 公開日 | 2013-06-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystallographic analysis and structure-guided engineering of NADPH-dependent Ralstonia sp. Alcohol dehydrogenase toward NADH cosubstrate specificity.
Biotechnol.Bioeng., 110, 2013
|
|
4H8I
 
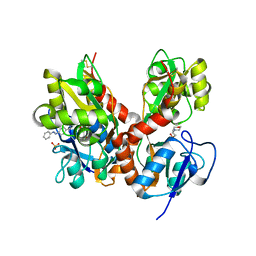 | | Structure of GluK2-LBD in complex with GluAzo | | 分子名称: | (4R)-4-[(2E)-3-{4-[(E)-phenyldiazenyl]phenyl}prop-2-en-1-yl]-L-glutamic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Reiter, A, Skerra, A, Trauner, D, Schiefner, A. | | 登録日 | 2012-09-22 | | 公開日 | 2013-09-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A photoswitchable neurotransmitter analogue bound to its receptor.
Biochemistry, 52, 2013
|
|
4GRX
 
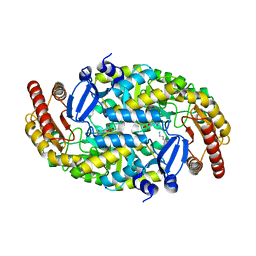 | | Structure of an omega-aminotransferase from Paracoccus denitrificans | | 分子名称: | Aminotransferase, DELTA-AMINO VALERIC ACID, SODIUM ION | | 著者 | Rausch, C, Lerchner, A, Schiefner, A, Skerra, A. | | 登録日 | 2012-08-27 | | 公開日 | 2012-12-19 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Crystal structure of the omega-aminotransferase from Paracoccus denitrificans and its phylogenetic relationship with other class III aminotransferases that have biotechnological potential.
Proteins, 81, 2013
|
|
4H8J
 
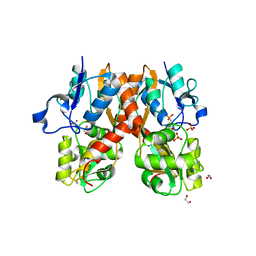 | | Structure of GluA2-LBD in complex with MES | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Glutamate receptor 2, ... | | 著者 | Reiter, A, Skerra, A, Trauner, D, Schiefner, A. | | 登録日 | 2012-09-22 | | 公開日 | 2013-09-25 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of an artificial photoreceptor
To be Published
|
|
3P7H
 
 | |
3P7G
 
 | |
3P7F
 
 | |
