1ZBB
 
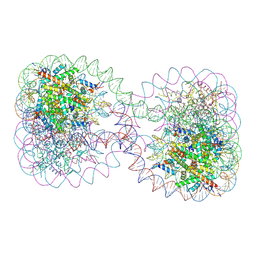 | | Structure of the 4_601_167 Tetranucleosome | | 分子名称: | DNA STRAND 1 (ARBITRARY MODEL SEQUENCE), DNA STRAND 2 (ARBITRARY MODEL SEQUENCE), HISTONE H3, ... | | 著者 | Schalch, T, Duda, S, Sargent, D.F, Richmond, T.J. | | 登録日 | 2005-04-08 | | 公開日 | 2005-07-12 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (9 Å) | | 主引用文献 | X-ray structure of a tetranucleosome and its implications for the chromatin fibre.
Nature, 436, 2005
|
|
3G7L
 
 | |
3TIX
 
 | |
6Z2A
 
 | |
5OY7
 
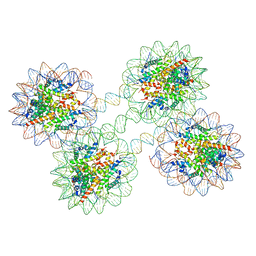 | | Structure of the 4_601_157 tetranucleosome (P1 form) | | 分子名称: | CHLORIDE ION, DNA (619-MER), Histone H2A, ... | | 著者 | Ekundayo, B, Richmond, T.J, Schalch, T. | | 登録日 | 2017-09-07 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (5.774 Å) | | 主引用文献 | Capturing Structural Heterogeneity in Chromatin Fibers.
J. Mol. Biol., 429, 2017
|
|
5OXV
 
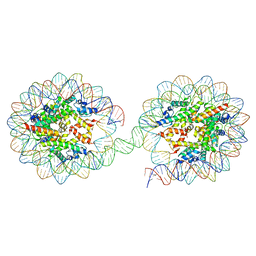 | |
4O9D
 
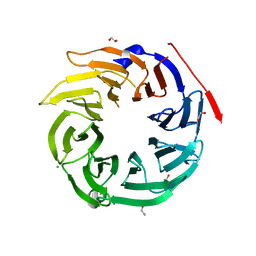 | | Structure of Dos1 propeller | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Rik1-associated factor 1 | | 著者 | Kuscu, C, Schalch, T, Joshua-Tor, L. | | 登録日 | 2014-01-02 | | 公開日 | 2014-01-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CRL4-like Clr4 complex in Schizosaccharomyces pombe depends on an exposed surface of Dos1 for heterochromatin silencing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5IKK
 
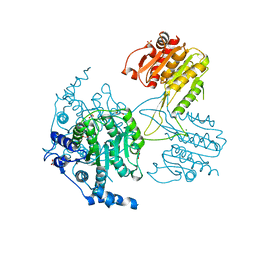 | |
5IKF
 
 | |
5IKJ
 
 | |
6EXT
 
 | |
6EXU
 
 | |
6FTO
 
 | |
