1UDI
 
 | |
1UDH
 
 | |
1UDG
 
 | |
4L5N
 
 | | Crystallographic Structure of HHV-1 Uracil-DNA Glycosylase complexed with the Bacillus phage PZA inhibitor protein p56 | | 分子名称: | ACETATE ION, Early protein GP1B, Uracil-DNA glycosylase | | 著者 | Cole, A.R, Sapir, O, Ryzhenkova, K, Baltulionis, G, Hornyak, P, Savva, R. | | 登録日 | 2013-06-11 | | 公開日 | 2013-08-07 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Architecturally diverse proteins converge on an analogous mechanism to inactivate Uracil-DNA glycosylase.
Nucleic Acids Res., 41, 2013
|
|
8AIN
 
 | | MCUGI SAUNG complex | | 分子名称: | 1,2-ETHANEDIOL, MCUGI, SULFATE ION, ... | | 著者 | Muselmani, W, Savva, R. | | 登録日 | 2022-07-26 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
8AIM
 
 | | Ugi-2 SAUNG complex | | 分子名称: | Uracil-DNA glycosylase, Uracil-DNA glycosylase inhibitor | | 著者 | Muselmani, W, Savva, R. | | 登録日 | 2022-07-26 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
8AIL
 
 | | Bacillus phage VMY22 p56 in complex with Bacillus weidmannii Ung | | 分子名称: | Bacillus phage VMY22 p56, GLYCEROL, IODIDE ION, ... | | 著者 | Muselmani, W, Bagneris, C, Savva, R. | | 登録日 | 2022-07-26 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | A Multimodal Approach towards Genomic Identification of Protein Inhibitors of Uracil-DNA Glycosylase.
Viruses, 15, 2023
|
|
2C53
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | 分子名称: | 2'-DEOXYURIDINE, GLYCEROL, URACIL DNA GLYCOSYLASE | | 著者 | Krusong, K, Carpenter, E.P, Bellmy, S.R.W, Savva, R, Baldwin, G.S. | | 登録日 | 2005-10-25 | | 公開日 | 2005-11-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2C56
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | 分子名称: | URACIL DNA GLYCOSYLASE, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Krusong, K, Carpenter, E.P, Bellamy, S.R.W, Savva, R, Baldwin, G.S. | | 登録日 | 2005-10-26 | | 公開日 | 2005-11-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
5NNH
 
 | | KSHV uracil-DNA glycosylase, apo form | | 分子名称: | SULFATE ION, Uracil-DNA glycosylase | | 著者 | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | 登録日 | 2017-04-09 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NN7
 
 | | KSHV uracil-DNA glycosylase, apo form | | 分子名称: | Uracil-DNA glycosylase | | 著者 | Earl, C, Bagneris, C, Cole, A.R, Barrett, T, Savva, R. | | 登録日 | 2017-04-08 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
5NNU
 
 | | KSHV uracil-DNA glycosylase, product complex with dsDNA exhibiting duplex nucleotide flipping | | 分子名称: | DNA, DNA containing an abasic site, Uracil-DNA glycosylase | | 著者 | Earl, C, Bagneris, C, Barrett, T, Savva, R. | | 登録日 | 2017-04-10 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | A structurally conserved motif in gamma-herpesvirus uracil-DNA glycosylases elicits duplex nucleotide-flipping.
Nucleic Acids Res., 46, 2018
|
|
1LAU
 
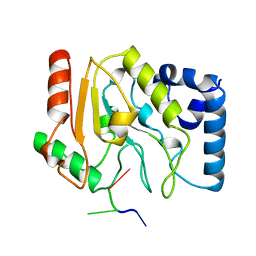 | | URACIL-DNA GLYCOSYLASE | | 分子名称: | DNA (5'-D(*TP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE (E.C.3.2.2.-)) | | 著者 | Pearl, L.H, Savva, R. | | 登録日 | 1996-01-03 | | 公開日 | 1996-06-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structural basis of specific base-excision repair by uracil-DNA glycosylase.
Nature, 373, 1995
|
|
7PV2
 
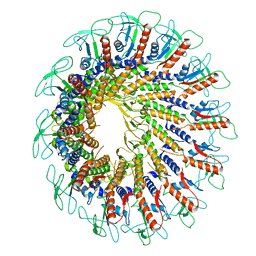 | |
7PV4
 
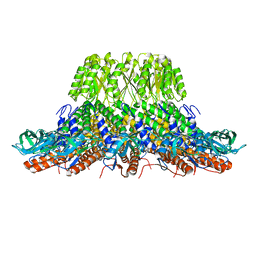 | |
1MWJ
 
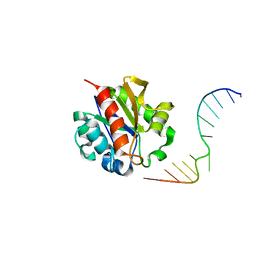 | | Crystal Structure of a MUG-DNA pseudo substrate complex | | 分子名称: | 5'-D(*CP*GP*CP*GP*A*GP*(DU)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | 著者 | Barrett, T.E, Scharer, O, Savva, R, Brown, T, Jiricny, J, Verdine, G.L, Pearl, L.H. | | 登録日 | 2002-09-30 | | 公開日 | 2002-10-11 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Crystal Structure of a thwarted mismatch glycosylase DNA repair complex
Embo J., 18, 1999
|
|
1MTL
 
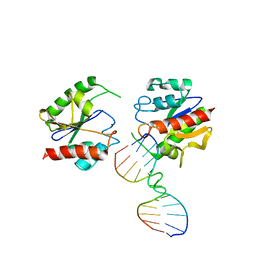 | | Non-productive MUG-DNA complex | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | 著者 | Barrett, T.E, Savva, R, Barlow, T, Brown, T, Jiricny, J, Pearl, L.H. | | 登録日 | 2002-09-21 | | 公開日 | 2002-09-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of a DNA base-excision product resembling a cisplatin inter-strand adduct.
Nat.Struct.Biol., 5, 1998
|
|
1MWI
 
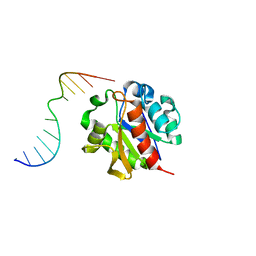 | | Crystal structure of a MUG-DNA product complex | | 分子名称: | 5'-D(*CP*GP*CP*GP*AP*GP*(AAB)P*TP*CP*GP*CP*G)-3', G/U mismatch-specific DNA glycosylase | | 著者 | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | 登録日 | 2002-09-30 | | 公開日 | 2002-10-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
1MUG
 
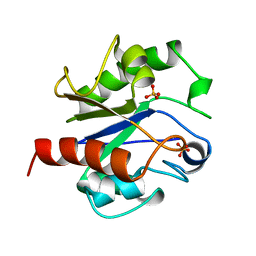 | | G:T/U MISMATCH-SPECIFIC DNA GLYCOSYLASE FROM E.COLI | | 分子名称: | PROTEIN (G:T/U SPECIFIC DNA GLYCOSYLASE), SULFATE ION | | 著者 | Barrett, T.E, Savva, R, Panayotou, G, Brown, T, Barlow, T, Jiricny, J, Pearl, L.H. | | 登録日 | 1998-07-10 | | 公開日 | 1998-07-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions.
Cell(Cambridge,Mass.), 92, 1998
|
|
1ODG
 
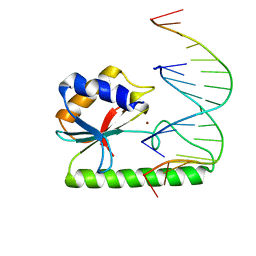 | | Very-short-patch DNA repair endonuclease bound to its reaction product site | | 分子名称: | 5'-D(*TP*AP*GP*GP*CP*5CM*TP*GP*GP*AP*TP*CP)-3', DNA MISMATCH ENDONUCLEASE, ZINC ION | | 著者 | Bunting, K.A, Roe, S.M, Headley, A, Brown, T, Savva, R, Pearl, L.H. | | 登録日 | 2003-02-19 | | 公開日 | 2003-03-13 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of the Escherichia Coli Dcm Very-Short-Patch DNA Repair Endonuclease Bound to its Reaction Product-Site in a DNA Superhelix
Nucleic Acids Res., 31, 2003
|
|
