3K8N
 
 | |
3NKA
 
 | | Crystal structure of AqpZ H174G,T183F | | Descriptor: | Aquaporin Z, GLYCEROL, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell III, J.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NKC
 
 | | Crystal structure of AqpZ F43W,H174G,T183F | | Descriptor: | Aquaporin Z, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell III, J.D, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 2010-06-18 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NK5
 
 | | Crystal structure of AqpZ mutant F43W | | Descriptor: | Aquaporin Z, octyl beta-D-glucopyranoside | | Authors: | Savage, D.F, O'Connell, J.D, Stroud, R.M, Finer-Moore, J.S. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural context shapes the aquaporin selectivity filter.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2O9F
 
 | |
2O9E
 
 | |
2O9G
 
 | |
2O9D
 
 | |
1RC2
 
 | | 2.5 Angstrom Resolution X-ray Structure of Aquaporin Z | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, Aquaporin Z | | Authors: | Savage, D.F, Egea, P.F, Robles, Y.C, O'Connell III, J.D, Stroud, R.M. | | Deposit date: | 2003-11-03 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z
Plos Biol., 1, 2003
|
|
7MU1
 
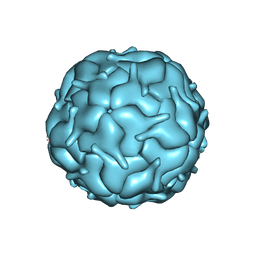 | | Thermotoga maritima encapsulin shell | | Descriptor: | FLAVIN MONONUCLEOTIDE, Maritimacin | | Authors: | LaFrance, B.J, Nogales, E, Savage, D.F. | | Deposit date: | 2021-05-14 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The encapsulin from Thermotoga maritima is a flavoprotein with a symmetry matched ferritin-like cargo protein.
Sci Rep, 11, 2021
|
|
7SNV
 
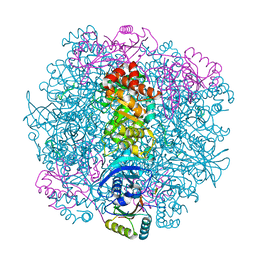 | | H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex | | Descriptor: | Carboxysome shell carbonic anhydrase, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Blikstad, C, Dugan, E, Laughlin, T.G, Liu, M, Shoemaker, S, Remis, J, Savage, D.F. | | Deposit date: | 2021-10-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Identification of a carbonic anhydrase-Rubisco complex within the alpha-carboxysome.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SMK
 
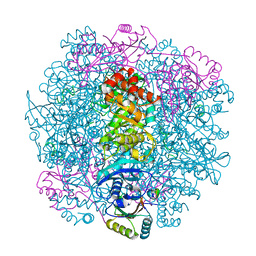 | | H. neapolitanus carboxysomal rubisco/CsoSCA-peptide (1-50)complex | | Descriptor: | Carboxysome shell carbonic anhydrase, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain | | Authors: | Blikstad, C, Dugan, E, Laughlin, T.G, Liu, M, Shoemaker, S, Remis, J, Savage, D.F. | | Deposit date: | 2021-10-26 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Identification of a carbonic anhydrase-Rubisco complex within the alpha-carboxysome.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6UEW
 
 | |
1NM2
 
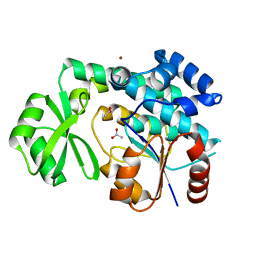 | | Malonyl-CoA:ACP Transacylase | | Descriptor: | ACETIC ACID, NICKEL (II) ION, malonyl CoA:acyl carrier protein malonyltransferase | | Authors: | Keatinge-Clay, A.T, Shelat, A.A, Savage, D.F, Tsai, S, Miercke, L.J.W, O'Connell III, J.D, Khosla, C, Stroud, R.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-01-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalysis, Specificity, and ACP Docking Site of
Streptomyces coelicolor Malonyl-CoA:ACP Transacylase
Structure, 11, 2003
|
|
6NBQ
 
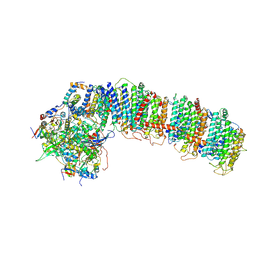 | | T.elongatus NDH (data-set 1) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 2, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBX
 
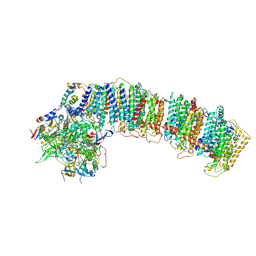 | | T.elongatus NDH (data-set 2) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6NBY
 
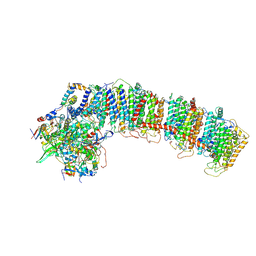 | | T.elongatus NDH (composite model) | | Descriptor: | IRON/SULFUR CLUSTER, NAD(P)H-quinone oxidoreductase chain 4 1, NAD(P)H-quinone oxidoreductase subunit 1, ... | | Authors: | Laughlin, T.G, Bayne, A, Trempe, J.-F, Savage, D.F, Davies, K.M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-02-27 | | Last modified: | 2020-04-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the complex I-like molecule NDH of oxygenic photosynthesis.
Nature, 566, 2019
|
|
6X8M
 
 | | CryoEM structure of the holo-SrpI encapsulin complex from Synechococcus elongatus PCC 7942 | | Descriptor: | Protein SrpI | | Authors: | LaFrance, B.J, Nichols, R.J, Phillips, N.R, Oltrogge, L.M, Valentin-Alvarado, L.E, Bischoff, A.J, Savage, D.F, Nogales, E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism.
Elife, 10, 2021
|
|
6X8T
 
 | | CryoEM structure of the apo-SrpI encapasulin complex from Synechococcus elongatus PCC 7942 | | Descriptor: | Protein SrpI | | Authors: | LaFrance, B.J, Nichols, R.J, Phillips, N.R, Oltrogge, L.M, Valentin-Alvarado, L.E, Bischoff, A.J, Savage, D.F, Nogales, E. | | Deposit date: | 2020-06-01 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery and characterization of a novel family of prokaryotic nanocompartments involved in sulfur metabolism.
Elife, 10, 2021
|
|
1RJ9
 
 | | Structure of the heterodimer of the conserved GTPase domains of the Signal Recognition Particle (Ffh) and Its Receptor (FtsY) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Signal Recognition Protein, ... | | Authors: | Egea, P.F, Shan, S.O, Napetschnig, J, Savage, D.F, Walter, P, Stroud, R.M. | | Deposit date: | 2003-11-18 | | Release date: | 2004-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate twinning activates the signal recognition particle and its receptor
Nature, 427, 2004
|
|
