4KC8
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from Thermotoga petrophila RKU-1 in complex with TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Glycoside hydrolase, ... | | Authors: | Nascimento, A.F.Z, Polo, C.C, Santos, C.R, Costa, M.C.M.F, Mesa, A.N, Prade, R.A, Ruller, R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
1FZS
 
 | | DNA WITH PYRENE PAIRED AT ABASIC SITE | | Descriptor: | 5'-D(*CP*AP*CP*AP*AP*AP*CP*AP*(PYP))-3', 5'-D(*GP*TP*GP*CP*(3DR)P*TP*GP*TP*TP*TP*GP*TP*G)-3' | | Authors: | Smirnov, S, Matray, T.J, Kool, E.T, de los Santos, C. | | Deposit date: | 2000-10-04 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Integrity of duplex structures without hydrogen bonding: DNA with pyrene paired at abasic sites
Nucleic Acids Res., 30, 2002
|
|
4NPR
 
 | | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase GH12 | | Authors: | Cordeiro, R.L, Santos, C.R, Furtado, G.P, Damasio, A.R.L, Polizeli, M.L.T.M, Ward, R.J, Murakami, M.T. | | Deposit date: | 2013-11-22 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Family 12 Xyloglucanase from Aspergillus niveus
To be Published
|
|
2LGM
 
 | | Structure of DNA Containing an Aristolactam II-dA Lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*AP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3'), [1,3]benzodioxolo[6,5,4-cd]benzo[f]indol-5(6H)-one | | Authors: | Lukin, M, Zaliznyak, T, Johnson, F, de los Santos, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and stability of DNA containing an aristolactam II-dA lesion: implications for the NER recognition of bulky adducts.
Nucleic Acids Res., 40, 2012
|
|
2LWO
 
 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*G*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
2LWN
 
 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
2LWM
 
 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
6UQJ
 
 | |
2GE2
 
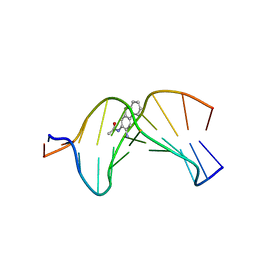 | |
2LT0
 
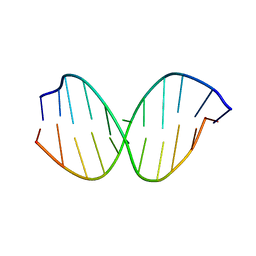 | | NMR structure of duplex DNA containing the beta-OH-PdG dA base pair: A mutagenic intermediate of acrolein | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(63G)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3') | | Authors: | Zaliznyak, T, de los Santos, C, Lukin, M, El-khateeb, M, Bonala, R, Johnson, F. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of duplex DNA containing the alpha-OH-PdG.dA base pair: a mutagenic intermediate of acrolein.
Biopolymers, 93, 2010
|
|
2KDA
 
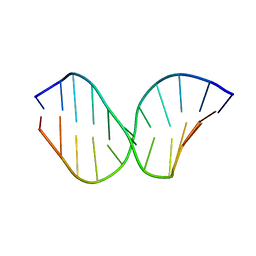 | | Solution Structure of DNA Containing Alpha-OH-PdG: the Mutagenic Adduct Produced by Acrolein | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(63H)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3' | | Authors: | de los Santos, C, Zaliznyak, T, Johnson, F, Bonala, R, Attaluri, S. | | Deposit date: | 2009-01-05 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of DNA containing alpha-OH-PdG: the mutagenic adduct produced by acrolein.
Nucleic Acids Res., 37, 2009
|
|
6UB1
 
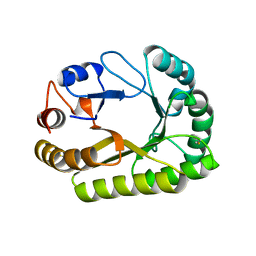 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -3 and -2 subsites | | Descriptor: | GLYCOSIDE HYDROLASE, beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAR
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAU
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaritriose and laminaribiose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glyco_hydro_cc domain-containing protein, ZINC ION, ... | | Authors: | Vieira, P.S, Cabral, L, Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UB0
 
 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -2 and -1 subsites | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAQ
 
 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | GLYCEROL, Glyco_hydro_cc domain-containing protein, SODIUM ION | | Authors: | Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAT
 
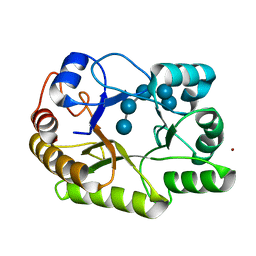 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E102A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose | | Descriptor: | Glyco_hydro_cc domain-containing protein, ZINC ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Vieira, P.S, Cabral, L, Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAY
 
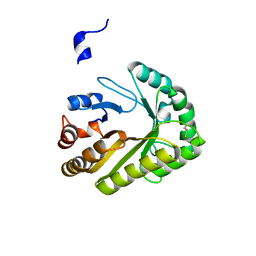 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) | | Descriptor: | GLYCOSIDE HYDROLASE | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAS
 
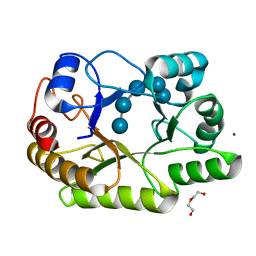 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199A mutant) from Amycolatopsis mediterranei (AmGH128_I) in complex with laminaripentaose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glycoside Hydrolase, ZINC ION, ... | | Authors: | Vieira, P.S, Cabral, L, Costa, P.A.C.R, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UFL
 
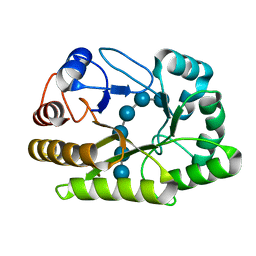 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) in the complex with laminarihexaose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose | | Authors: | Cordeiro, R.L, Domingues, M.N, Vieira, P.S, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-24 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UAZ
 
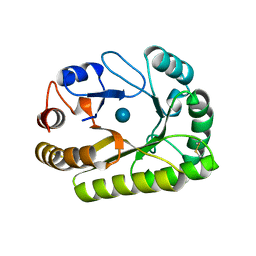 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with glucose | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UFZ
 
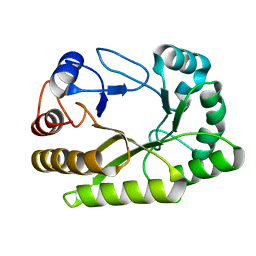 | | Crystal structure of a GH128 (subgroup I) endo-beta-1,3-glucanase (E199Q mutant) from Amycolatopsis mediterranei (AmGH128_I) | | Descriptor: | Glyco_hydro_cc domain-containing protein | | Authors: | Cordeiro, R.L, Domingues, M.N, Vieira, P.S, Santos, C.R, Murakami, M.T. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
2K1Y
 
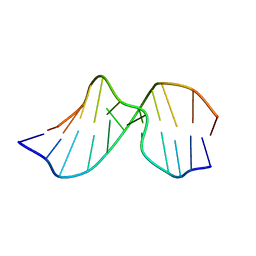 | | Solution Structure of Duplex DNA Containing the Mutagenic Lesion: 1,N2-Etheno-2'-deoxyguanine | | Descriptor: | 5'-D(*DCP*DGP*DTP*DAP*DCP*(GNE)P*DCP*DAP*DTP*DGP*DC)-3', 5'-D(*DGP*DCP*DAP*DTP*DGP*DCP*DGP*DTP*DAP*DCP*DG)-3' | | Authors: | Zaliznyak, T, Lukin, M, Johnson, F, de los Santos, C. | | Deposit date: | 2008-03-18 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of duplex DNA containing the mutagenic lesion 1,N(2)-etheno-2'-deoxyguanine.
Biochemistry, 47, 2008
|
|
3PRV
 
 | | Nucleoside diphosphate kinase B from Trypanosoma cruzi | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Souza, T.A.C.B, Trindade, D.M, Tonoli, C.C.C, Santos, C.R, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-11-30 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
3NGS
 
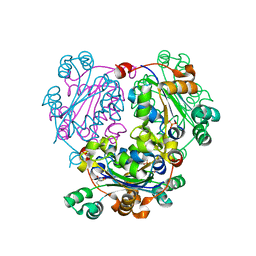 | | Structure of Leishmania nucleoside diphosphate kinase b with ordered nucleotide-binding loop | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Nucleoside diphosphate kinase, PHOSPHATE ION | | Authors: | Trindade, D.M, Sousa, T.A.C.B, Tonoli, C.C.C, Santos, C.R, Arni, R.K, Ward, R.J, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-06-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
