7QNO
 
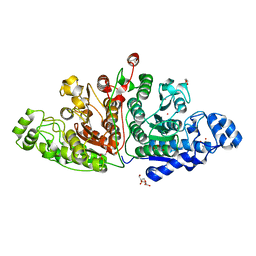 | | Crystal structure of ligand-free Danio rerio HDAC6 CD1 CD2 | | Descriptor: | GLYCEROL, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Kempf, G, Langousis, G, Sanchez, J, Matthias, P. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Expression and Crystallization of HDAC6 Tandem Catalytic Domains.
Methods Mol.Biol., 2589, 2023
|
|
1OFZ
 
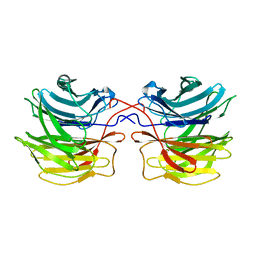 | | Crystal structure of fungal lectin : six-bladed beta-propeller fold and novel fucose recognition mode for aleuria aurantia lectin | | Descriptor: | FUCOSE-SPECIFIC LECTIN, alpha-L-fucopyranose, beta-L-fucopyranose | | Authors: | Wimmerova, M, Mitchell, E, Sanchez, J.F, Gautier, C, Imberty, A. | | Deposit date: | 2003-04-22 | | Release date: | 2003-07-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Fungal Lectin: Six-Bladed {Beta}-Propeller Fold and Novel Fucose Recognition Mode for Aleuria Aurantia Lectin
J.Biol.Chem., 278, 2003
|
|
4LTB
 
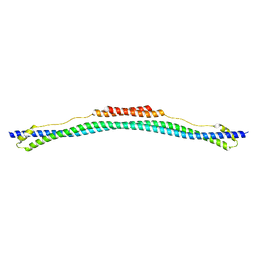 | | Coiled-coil domain of TRIM25 | | Descriptor: | Tripartite motif-containing 25 variant | | Authors: | Pornillos, O, Sanchez, J.G, Okreglicka, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5EYA
 
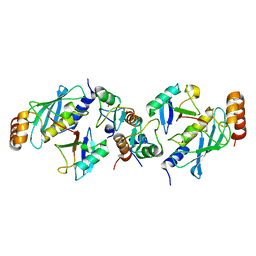 | | TRIM25 RING domain in complex with Ubc13-Ub conjugate | | Descriptor: | Polyubiquitin-B, Tripartite motif-containing 25 variant, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Pornillos, O, Sanchez, J.G. | | Deposit date: | 2015-11-24 | | Release date: | 2016-08-17 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mechanism of TRIM25 Catalytic Activation in the Antiviral RIG-I Pathway.
Cell Rep, 16, 2016
|
|
1N5H
 
 | | Solution structure of the cathelin-like domain of protegrins (the R87-P88 and D118-P119 amide bonds are in the cis conformation) | | Descriptor: | protegrins | | Authors: | Yang, Y, Sanchez, J.F, Strub, M.P, Brutscher, B, Aumelas, A. | | Deposit date: | 2002-11-06 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Cathelin-like domain of the protegrin-3 Precursor
Biochemistry, 42, 2003
|
|
1N5P
 
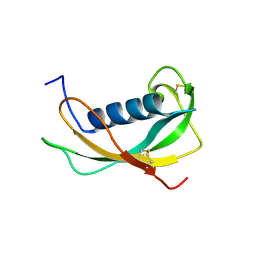 | | Solution structure of the cathelin-like domain of protegrins (all amide bonds involving proline residues are in trans conformation) | | Descriptor: | protegrins | | Authors: | Yang, Y, Sanchez, J.F, Strub, M.P, Brutscher, B, Aumelas, A. | | Deposit date: | 2002-11-07 | | Release date: | 2003-06-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Cathelin-like domain of the protegrin-3 Precursor
Biochemistry, 42, 2003
|
|
5G4I
 
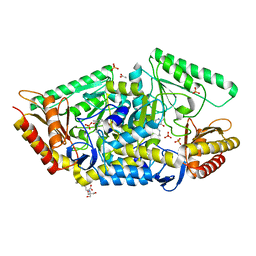 | | PLP-dependent phospholyase A1RDF1 from Arthrobacter aurescens TC1 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
5G4J
 
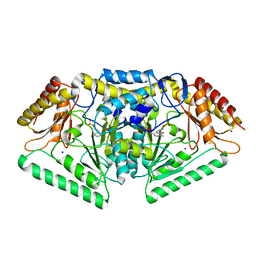 | | Phospholyase A1RDF1 from Arthrobacter in complex with phosphoethanolamine | | Descriptor: | PUTATIVE AMINOTRANSFERASE CLASS III PROTEIN, SODIUM ION, {5-hydroxy-6-methyl-4-[(E)-{[2-(phosphonooxy)ethyl]imino}methyl]pyridin-3-yl}methyl dihydrogen phosphate | | Authors: | Cuetos, A, Tuan, A.N, Mangas Sanchez, J, Grogan, G. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for Phospholyase Activity of a Class III Transaminase Homologue.
Chembiochem, 17, 2016
|
|
