2J7O
 
 | | STRUCTURE OF THE RNAI POLYMERASE FROM NEUROSPORA CRASSA | | Descriptor: | MAGNESIUM ION, RNA DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
4AOM
 
 | | MTIP and MyoA complex | | Descriptor: | MYOSIN A TAIL DOMAIN INTERACTING PROTEIN, MYOSIN-A | | Authors: | Salgado, P.S, Douse, C.H, Simpson, P.J, Thomas, J.C, Holder, A.A, Tate, E.W, Cota, E. | | Deposit date: | 2012-03-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Regulation of the Plasmodium Motor Complex: Phosphorylation of Myosin a Tail Interacting Protein (Mtip) Loosens its Grip on Myoa
J.Biol.Chem., 287, 2012
|
|
2J7N
 
 | | Structure of the RNAi polymerase from Neurospora crassa | | Descriptor: | GLYCEROL, MAGNESIUM ION, RNA-DEPENDENT RNA POLYMERASE | | Authors: | Salgado, P.S, Koivunen, M.R.L, Makeyev, E.V, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of an Rnai Polymerase Links RNA Silencing and Transcription.
Plos Biol., 4, 2006
|
|
2Y7M
 
 | |
2Y7O
 
 | |
2Y7N
 
 | |
2XSK
 
 | | E. coli curli protein CsgC - SeCys | | Descriptor: | ACETATE ION, CSGC | | Authors: | Salgado, P.S, Taylor, J.D, Cota, E, Matthews, S.J. | | Deposit date: | 2010-09-29 | | Release date: | 2010-12-29 | | Last modified: | 2014-01-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Extending the Usability of the Phasing Power of Diselenide Bonds: Secys Sad Phasing of Csgc Using a Non-Auxotrophic Strain.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2Y7L
 
 | |
2YLH
 
 | |
1UVJ
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 7nt RNA | | Descriptor: | 5'-R(*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVN
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 ca2+ inhibition complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*CP*CP)-3', CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVL
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5nt RNA. Conformation B | | Descriptor: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVI
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 6nt RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVK
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 dead-end complex | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVM
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5NT RNA conformation A | | Descriptor: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
8BBY
 
 | |
7ACZ
 
 | |
7ACV
 
 | |
7ACY
 
 | |
7ACX
 
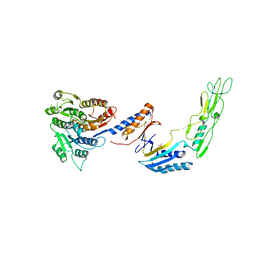 | | H/L (SLPH/SLPL) complex from C. difficile (R7404 strain) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-layer protein, SULFATE ION | | Authors: | Lanzoni-Mangutchi, P, Barwinska-Sendra, A, Basle, A, El Omari, K, Wagner, A, Salgado, P.S. | | Deposit date: | 2020-09-11 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and assembly of the S-layer in C. difficile.
Nat Commun, 13, 2022
|
|
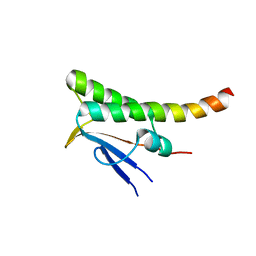
7ACW
 
 | |
2JLF
 
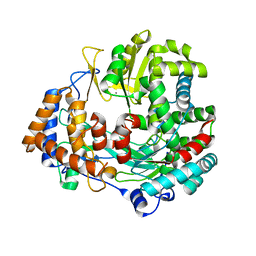 | | STRUCTURAL EXPLANATION FOR THE ROLE OF MN IN THE ACTIVITY OF PHI6 RNA- DEPENDENT RNA POLYMERASE | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-08 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2JLG
 
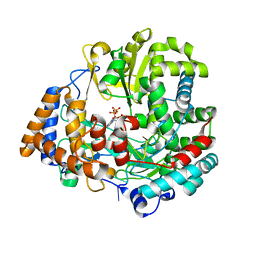 | | STRUCTURAL EXPLANATION FOR THE ROLE OF MN IN THE ACTIVITY OF PHI6 RNA-DEPENDENT RNA POLYMERASE | | Descriptor: | 5'-D(*DT DT DT DC DCP)-3', GUANOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-09 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2JL9
 
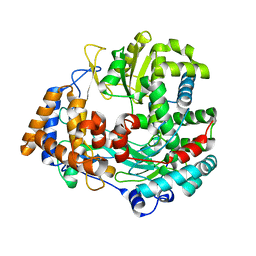 | | Structural explanation for the role of Mn in the activity of phi6 RNA- dependent RNA polymerase | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Poranen, M.M, Salgado, P.S, Koivunen, M.R.L, Wright, S, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2008-09-05 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Explanation for the Role of Mn2+ in the Activity of {Phi}6 RNA-Dependent RNA Polymerase.
Nucleic Acids Res., 36, 2008
|
|
2Y2T
 
 | |
