8XEJ
 
 | | Cryo-EM structure of human XKR8-basigin complex in lipid nanodisc | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab heavy chain, Fab light chain, ... | | Authors: | Sakuragi, T.S, Kanai, R.K, Kikkawa, M.K, Toyoshima, C.T, Nagata, S.N. | | Deposit date: | 2023-12-12 | | Release date: | 2024-02-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | The role of the C-terminal tail region as a plug to regulate XKR8 lipid scramblase.
J.Biol.Chem., 300, 2024
|
|
7DAA
 
 | | Crystal structure of basigin complexed with anti-basigin Fab fragment | | Descriptor: | CADMIUM ION, Heavy chain of antibody Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7DCE
 
 | | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Heavy chain of Fab fragment, Isoform 2 of Basigin, ... | | Authors: | Sakuragi, T, Kanai, R, Tsutsumi, A, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Kikkawa, M, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-26 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D9Z
 
 | | Crystal structure of anti-basigin Fab fragment | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Heavy chain of antibody Fab fragment, ... | | Authors: | Sakuragi, T, Kanai, R, Narita, H, Onishi, E, Miyazaki, T, Baba, T, Nakagawa, A, Toyoshima, C, Nagata, S. | | Deposit date: | 2020-10-14 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.123 Å) | | Cite: | The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1IUP
 
 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant complexed with isobutyrates | | Descriptor: | 2-METHYL-PROPIONIC ACID, meta-Cleavage product hydrolase | | Authors: | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
1IUN
 
 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant hexagonal | | Descriptor: | ACETATE ION, meta-Cleavage product hydrolase | | Authors: | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
1IUO
 
 | | meta-Cleavage product hydrolase from Pseudomonas fluorescens IP01 (CumD) S103A mutant complexed with acetates | | Descriptor: | ACETATE ION, meta-Cleavage product hydrolase | | Authors: | Fushinobu, S, Saku, T, Hidaka, M, Jun, S.-Y, Nojiri, H, Yamane, H, Shoun, H, Omori, T, Wakagi, T. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a meta-cleavage product hydrolase from Pseudomonas
fluorescens IP01 (CumD) complexed with cleavage products
PROTEIN SCI., 11, 2002
|
|
1D7N
 
 | | SOLUTION STRUCTURE ANALYSIS OF THE MASTOPARAN WITH DETERGENTS | | Descriptor: | PROTEIN (WASP VENOM PEPTIDE (MASTOPARAN)) | | Authors: | Hori, Y, Demura, M, Iwadate, M, Niidome, T, Aoyagi, H, Asakura, T. | | Deposit date: | 1999-10-19 | | Release date: | 2001-06-20 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Interaction of mastoparan with membranes studied by 1H-NMR spectroscopy in detergent micelles and by solid-state 2H-NMR and 15N-NMR spectroscopy in oriented lipid bilayers.
Eur.J.Biochem., 268, 2001
|
|
7F19
 
 | | Structure of the G304E mutant of CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Takata, S.T, Kawano, T.K, Nakata, S.N, Takado, K.T, Imaizumi, R.I, Sakai, N.S, Takeshita, K.T, Yamashita, S.Y, Sakurai, T.S, Kataoka, K.K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure of the G304E mutant of CueO
To Be Published
|
|
1QXE
 
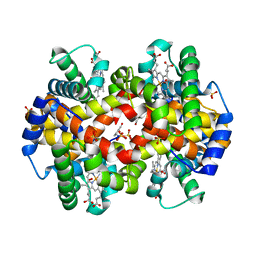 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | 5-HYDROXYMETHYL-FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
2Y44
 
 | | Crystal structure of GARP from Trypanosoma congolense | | Descriptor: | GLUTAMIC ACID/ALANINE-RICH PROTEIN, GLYCEROL, IODIDE ION | | Authors: | Loveless, B.C, Mason, J.W, Sakurai, T, Inoue, N, Razavi, M, Pearson, T.W, Boulanger, M.J. | | Deposit date: | 2011-01-04 | | Release date: | 2011-03-30 | | Last modified: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization and Epitope Mapping of the Glutamic Acid/Alanine-Rich Protein from Trypanosoma Congolense: Defining Assembly on the Parasite Cell Surface.
J.Biol.Chem., 286, 2011
|
|
7F0V
 
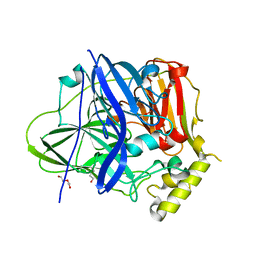 | | Structure of the M305I mutant of CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Kawano, T.K, Takata, S.T, Sakai, N.S, Imaizumi, R.I, Nakata, S.N, Takeshita, K.T, Yamashita, S.Y, Sakurai, T.S, Kataoka, K.K. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | Structure of the M305I mutant of CueO
To Be Published
|
|
1QXD
 
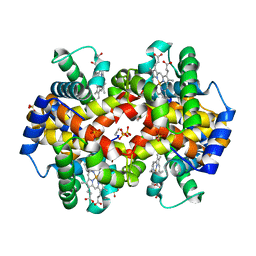 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
3LZX
 
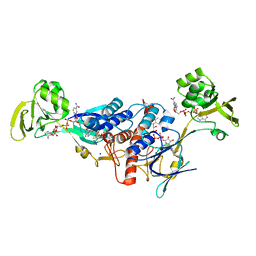 | | Crystal structure of ferredoxin-NADP+ oxidoreductase from Bacillus subtilis (FORM II) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Komori, H, Seo, D, Sakurai, T, Higuchi, Y. | | Deposit date: | 2010-03-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity
Protein Sci., 19, 2010
|
|
3LZW
 
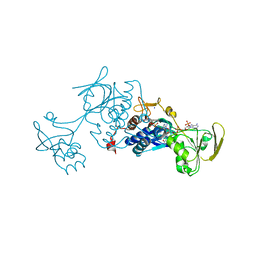 | | Crystal structure of ferredoxin-NADP+ oxidoreductase from bacillus subtilis (form I) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase 2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Komori, H, Seo, D, Sakurai, T, Higuchi, Y. | | Deposit date: | 2010-03-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure analysis of Bacillus subtilis ferredoxin-NADP(+) oxidoreductase and the structural basis for its substrate selectivity
Protein Sci., 19, 2010
|
|
1CCR
 
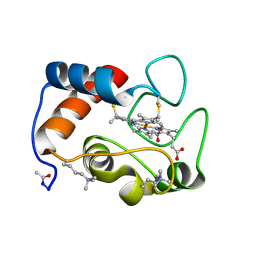 | | STRUCTURE OF RICE FERRICYTOCHROME C AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Ochi, H, Hata, Y, Tanaka, N, Kakudo, M, Sakurai, T, Aihara, S, Morita, Y. | | Deposit date: | 1983-03-14 | | Release date: | 1983-04-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of rice ferricytochrome c at 2.0 A resolution.
J.Mol.Biol., 166, 1983
|
|
2D04
 
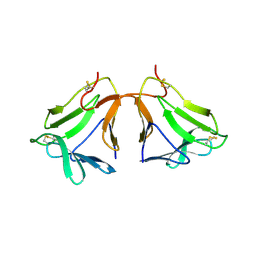 | | Crystal structure of neoculin, a sweet protein with taste-modifying activity. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Curculin, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimizu-Ibuka, A, Morita, Y, Terada, T, Asakura, T, Nakajima, K, Iwata, S, Misaka, T, Sorimachi, H, Arai, S, Abe, K. | | Deposit date: | 2005-07-25 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of neoculin: insights into its sweetness and taste-modifying activity
J.Mol.Biol., 359, 2006
|
|
4HAL
 
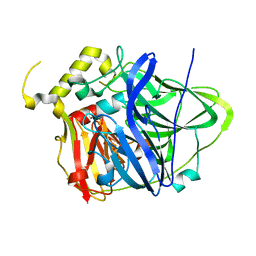 | | Multicopper Oxidase CueO mutant E506I | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multicopper Oxidase CueO mutant E506I
TO BE PUBLISHED
|
|
4HAK
 
 | | Multicopper Oxidase CueO mutant E506A | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Multicopper Oxidase CueO mutant E506A
TO BE PUBLISHED
|
|
4NER
 
 | | Multicopper Oxidase CueO (data1) | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION, HYDROXIDE ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2013-10-30 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New insights into the catalytic active-site structure of multicopper oxidases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3UAC
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data4) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UAA
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data1) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UAE
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data6) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UAD
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data5) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3UAB
 
 | | Multicopper Oxidase CueO mutant C500SE506Q (data2) | | Descriptor: | ACETATE ION, Blue copper oxidase CueO, COPPER (II) ION, ... | | Authors: | Komori, H, Kataoka, K, Sakurai, T, Higuchi, Y. | | Deposit date: | 2011-10-21 | | Release date: | 2012-04-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | An O-centered structure of the trinuclear copper center in the Cys500Ser/Glu506Gln mutant of CueO and structural changes in low to high X-ray dose conditions.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
