3H1Z
 
 | | Molecular basis for the association of PIPKIgamma -p90 with the clathrin adaptor AP-2 | | Descriptor: | AP-2 complex subunit beta-1, Phosphatidylinositol-4-phosphate 5-kinase type-1 gamma | | Authors: | Vahedi-Faridi, A, Kahlfeldt, N, Schaefer, J.G, Krainer, G, Keller, S, Saenger, W, Krauss, M, Haucke, V. | | Deposit date: | 2009-04-14 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular basis for association of PIPKI gamma-p90 with clathrin adaptor AP-2.
J.Biol.Chem., 285, 2010
|
|
1O7A
 
 | | Human beta-Hexosaminidase B | | Descriptor: | 1,2-ETHANEDIOL, 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maier, T, Strater, N, Schuette, C, Klingenstein, R, Sandhoff, K, Saenger, W. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2020-11-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The X-Ray Crystal Structure of Human Beta-Hexosaminidase B Provides New Insights Into Sandhoff Disease
J.Mol.Biol., 328, 2003
|
|
3H20
 
 | | Crystal structure of primase RepB' | | Descriptor: | DIPHOSPHATE, Replication protein B, SULFATE ION | | Authors: | Geibel, S, Banchenko, S, Engel, M, Lanka, E, Saenger, W. | | Deposit date: | 2009-04-14 | | Release date: | 2009-04-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure and function of primase RepB' encoded by broad-host-range plasmid RSF1010 that replicates exclusively in leading-strand mode
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GXV
 
 | | Three-dimensional structure of N-terminal domain of DnaB Helicase from Helicobacter pylori and its interactions with primase | | Descriptor: | Replicative DNA helicase | | Authors: | Kashav, T, Nitharwal, R, Syed, A.A, Gabdoulkhakov, A, Saenger, W, Dhar, K.S, Gourinath, S. | | Deposit date: | 2009-04-03 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of N-terminal domain of DnaB helicase and helicase-primase interactions in Helicobacter pylori
Plos One, 4, 2009
|
|
3HCV
 
 | | Crystal structure of HLA-B*2709 complexed with the double citrullinated vasoactive intestinal peptide type 1 receptor (VIPR) peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, DOUBLE CITRULLINATED VASOACTIVE INTESTINAL POLYPEPTIDE RECEPTOR, HLA class I histocompatibility antigen, ... | | Authors: | Beltrami, A, Gabdulkhakov, A, Rossmann, M, Ziegler, A, Uchanska-Ziegler, B, Saenger, W. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Citrullination-and mhc polymorphism-dependent conformational changes of
a self peptide
To be Published
|
|
3HS8
 
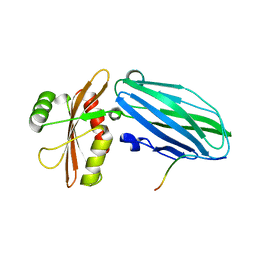 | | Intersectin 1-peptide-AP2 alpha ear complex | | Descriptor: | Adaptor protein complex AP-2, alpha 2 subunit, peptide from Intersectin-1, ... | | Authors: | Vahedi-Faridi, A, Pechstein, A, Schaefer, J.G, Saenger, W, Haucke, V. | | Deposit date: | 2009-06-10 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3HS9
 
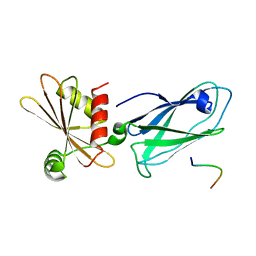 | | Intersectin 1-peptide-AP2 beta ear complex | | Descriptor: | AP-2 complex subunit beta-1, peptide from Intersectin-1, residues 841-851 | | Authors: | Vahedi-Faridi, A, Pechstein, A, Schaefer, J.G, Saenger, W, Haucke, V. | | Deposit date: | 2009-06-10 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Regulation of synaptic vesicle recycling by complex formation between intersectin 1 and the clathrin adaptor complex AP2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1PEK
 
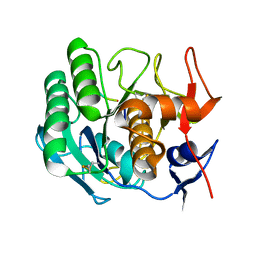 | | STRUCTURE OF THE COMPLEX OF PROTEINASE K WITH A SUBSTRATE-ANALOGUE HEXA-PEPTIDE INHIBITOR AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | D-DAL-ALA-NH2, PEPTIDE PRO-ALA-PRO-PHE, PROTEINASE K | | Authors: | Betzel, C, Singh, T.P, Visanji, M, Peters, K, Fittkau, S, Saenger, W, Wilson, K.S. | | Deposit date: | 1993-01-19 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the complex of proteinase K with a substrate analogue hexapeptide inhibitor at 2.2-A resolution.
J.Biol.Chem., 268, 1993
|
|
1QE5
 
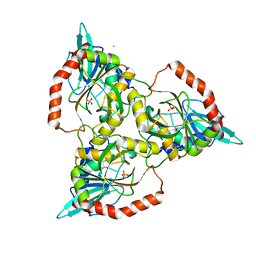 | | PURINE NUCLEOSIDE PHOSPHORYLASE FROM CELLULOMONAS SP. IN COMPLEX WITH PHOSPHATE | | Descriptor: | CALCIUM ION, PENTOSYLTRANSFERASE, PHOSPHATE ION | | Authors: | Tebbe, J, Bzowska, A, Wielgus-Kutrowska, B, Schroeder, W, Kazimierczuk, Z, Shugar, D, Saenger, W, Koellner, G. | | Deposit date: | 1999-07-13 | | Release date: | 1999-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the purine nucleoside phosphorylase (PNP) from Cellulomonas sp. and its implication for the mechanism of trimeric PNPs.
J.Mol.Biol., 294, 1999
|
|
2A83
 
 | | Crystal structure of hla-b*2705 complexed with the glucagon receptor (gr) peptide (residues 412-420) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Ruckert, C, Fiorillo, M.T, Loll, B, Moretti, R, Biesiadka, J, Saenger, W, Ziegler, A, Sorrentino, R, Uchanska-Ziegler, B. | | Deposit date: | 2005-07-07 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational dimorphism of self-peptides and molecular mimicry in a disease-associated HLA-B27 subtype.
J.Biol.Chem., 281, 2006
|
|
1UXS
 
 | | CRYSTAL STRUCTURE OF HLA-B*2705 COMPLEXED WITH THE LATENT MEMBRANE PROTEIN 2 PEPTIDE (LMP2)OF EPSTEIN-BARR VIRUS | | Descriptor: | BETA-2-MICROGLOBULIN, GENE TERMINAL PROTEIN (MEMBRANE PROTEIN LMP-2A/LMP-2B), GLYCEROL, ... | | Authors: | Hulsmeyer, M, Kozerski, C, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2004-03-01 | | Release date: | 2004-11-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Allele-Dependent Similarity between Viral and Self-Peptide Presentation by Hla-B27 Subtypes
J.Biol.Chem., 280, 2005
|
|
2TRT
 
 | | TETRACYCLINE REPRESSOR CLASS D | | Descriptor: | MAGNESIUM ION, TETRACYCLINE, TETRACYCLINE REPRESSOR CLASS D | | Authors: | Hinrichs, W, Kisker, C, Saenger, W. | | Deposit date: | 1994-03-04 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance.
Science, 264, 1994
|
|
2YI1
 
 | | Crystal structure of N-Acetylmannosamine kinase in complex with N- acetyl mannosamine 6-phosphate and ADP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
2YHW
 
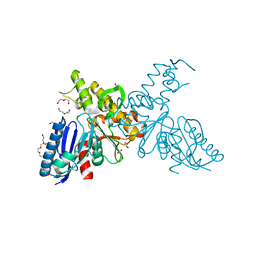 | | High-resolution crystal structures of N-Acetylmannosamine kinase: Insights about substrate specificity, activity and inhibitor modelling. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ACETATE ION, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Zimmer, R, Tauberger, E, Reutter, W, Saenger, W, Fan, H, Moniot, S. | | Deposit date: | 2011-05-08 | | Release date: | 2012-02-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
2YHY
 
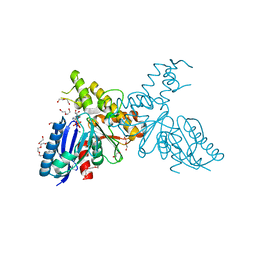 | | Structure of N-Acetylmannosamine kinase in complex with N- acetylmannosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-mannopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Martinez, J, Nguyen, L.D, Tauberger, E, Hinderlich, S, Reutter, W, Fan, H, Saenger, W, Moniot, S. | | Deposit date: | 2011-05-10 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures of N-Acetylmannosamine Kinase Provide Insights Into Enzyme Specificity and Inhibition
J.Biol.Chem., 287, 2012
|
|
1Q9E
 
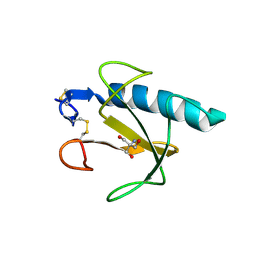 | | RNase T1 variant with adenine specificity | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Guanyl-specific ribonuclease T1 precursor | | Authors: | Czaja, R, Struhalla, M, Hoeschler, K, Saenger, W, Straeter, N, Hahn, U. | | Deposit date: | 2003-08-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNase T1 Variant RV Cleaves Single-Stranded RNA after Purines Due to Specific Recognition by the Asn46 Side Chain Amide.
Biochemistry, 43, 2004
|
|
1GVN
 
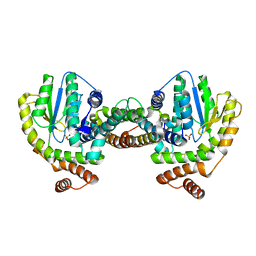 | | Crystal Structure of the Plasmid Maintenance System epsilon/zeta: Meachnism of toxin inactivation and toxin function | | Descriptor: | EPSILON, SULFATE ION, ZETA | | Authors: | Meinhart, A, Alonso, J.C, Straeter, N, Saenger, W. | | Deposit date: | 2002-02-19 | | Release date: | 2003-01-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Plasmid Maintenance System Epsilon /Zeta : Functional Mechanism of Toxin Zeta and Inactivation by Epsilon 2 Zeta 2 Complex Formation
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1RGC
 
 | | THE COMPLEX BETWEEN RIBONUCLEASE T1 AND 3'-GUANYLIC ACID SUGGESTS GEOMETRY OF ENZYMATIC REACTION PATH. AN X-RAY STUDY | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Heydenreich, A, Koellner, G, Choe, H.W, Cordes, F, Kisker, C, Schindelin, H, Adamiak, R, Hahn, U, Saenger, W. | | Deposit date: | 1993-05-12 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The complex between ribonuclease T1 and 3'GMP suggests geometry of enzymic reaction path. An X-ray study.
Eur.J.Biochem., 218, 1993
|
|
1IRQ
 
 | | Crystal structure of omega transcriptional repressor at 1.5A resolution | | Descriptor: | omega transcriptional repressor | | Authors: | Murayama, K, Orth, P, De La Hoz, A.B, Alonso, J.C, Saenger, W. | | Deposit date: | 2001-10-11 | | Release date: | 2001-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 A resolution.
J.Mol.Biol., 314, 2001
|
|
2BJG
 
 | | Crystal Structure of Conjugated Bile Acid Hydrolase from Clostridium perfringens in Complex with Reaction Products Taurine and Deoxycholate | | Descriptor: | 1,2-ETHANEDIOL, CHOLOYLGLYCINE HYDROLASE | | Authors: | Rossocha, M, Schultz-Heienbrok, R, Von Moeller, H, Coleman, J.P, Saenger, W. | | Deposit date: | 2005-02-02 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl But not of its Tauryl Product
Biochemistry, 44, 2005
|
|
2BJF
 
 | | Crystal Structure of Conjugated Bile Acid Hydrolase from Clostridium perfringens in Complex with Reaction Products Taurine and Deoxycholate | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, 2-AMINOETHANESULFONIC ACID, CHOLOYLGLYCINE HYDROLASE, ... | | Authors: | Rossocha, M, Schultz-Heienbrok, R, Von Moeller, H, Coleman, J.P, Saenger, W. | | Deposit date: | 2005-02-02 | | Release date: | 2005-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conjugated Bile Acid Hydrolase is a Tetrameric N-Terminal Thiol Hydrolase with Specific Recognition of its Cholyl But not of its Tauryl Product
Biochemistry, 44, 2005
|
|
2XZG
 
 | | Clathrin Terminal Domain Complexed with Pitstop 1 | | Descriptor: | 2-(4-AMINOBENZYL)-1,3-DIOXO-2,3-DIHYDRO-1H-BENZO[DE]ISOQUINOLINE-5-SULFONATE, ACETATE ION, CLATHRIN HEAVY CHAIN 1, ... | | Authors: | Bulut, H, Von Kleist, L, Saenger, W, Haucke, V. | | Deposit date: | 2010-11-25 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of the Clathrin Terminal Domain in Regulating Coated Pit Dynamics Revealed by Small Molecule Inhibition.
Cell(Cambridge,Mass.), 146, 2011
|
|
2HSG
 
 | |
1A69
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE IN COMPLEX WITH FORMYCIN B AND SULPHATE (PHOSPHATE) | | Descriptor: | FORMYCIN B, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Koellner, G, Luic, M, Shugar, D, Saenger, W, Bzowska, A. | | Deposit date: | 1998-03-08 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ternary complex of E. coli purine nucleoside phosphorylase with formycin B, a structural analogue of the substrate inosine, and phosphate (Sulphate) at 2.1 A resolution.
J.Mol.Biol., 280, 1998
|
|
2OZE
 
 | | The Crystal structure of Delta protein of pSM19035 from Streptoccocus pyogenes | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, Orf delta', ... | | Authors: | Cicek, A, Weihofen, W, Saenger, W. | | Deposit date: | 2007-02-26 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation.
Nucleic Acids Res., 36, 2008
|
|
