4LXC
 
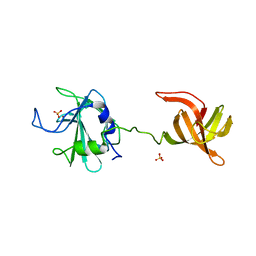 | | The antimicrobial peptidase lysostaphin from Staphylococcus simulans | | Descriptor: | Lysostaphin, SULFATE ION, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2013-07-29 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4QPB
 
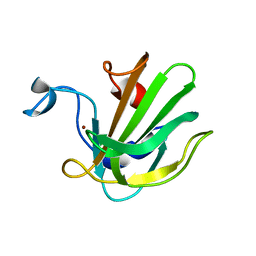 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the absence of phosphate | | Descriptor: | 1,2-ETHANEDIOL, Lysostaphin, ZINC ION | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
4QP5
 
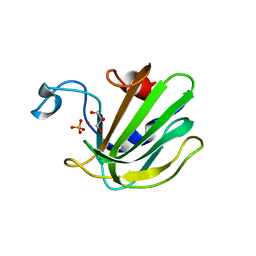 | | Catalytic domain of the antimicrobial peptidase lysostaphin from Staphylococcus simulans crystallized in the presence of phosphate | | Descriptor: | GLYCEROL, Lysostaphin, PHOSPHATE ION, ... | | Authors: | Sabala, I, Jagielska, E, Bardelang, P.T, Czapinska, H, Dahms, S.O, Sharpe, J.A, James, R, Than, M.E, Thomas, N.R, Bochtler, M. | | Deposit date: | 2014-06-22 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Crystal structure of the antimicrobial peptidase lysostaphin from Staphylococcus simulans.
Febs J., 281, 2014
|
|
6C68
 
 | | MHC-independent t cell receptor A11 | | Descriptor: | T-cell receptor alpha chain, T-cell receptor beta chain | | Authors: | Lu, J, Van Laethem, F, Saba, I, Chu, J, Bhattacharya, A, Love, N.C, Tikhonova, A, Radaev, S, Sun, X, Ko, A, Arnon, T, Shifrut, E, Friedman, N, Weng, N, Singer, A, Sun, P.D. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of MHC-Independent TCRs and Their Recognition of Native Antigen CD155.
J Immunol., 204, 2020
|
|
1TZP
 
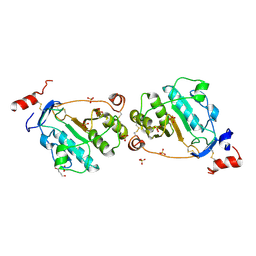 | | MEPA, inactive form without ZN in P21 | | Descriptor: | 1,4-BUTANEDIOL, Penicillin-insensitive murein endopeptidase, SULFATE ION | | Authors: | Marcyjaniak, M, Odintsov, S.G, Sabala, I, Bochtler, M. | | Deposit date: | 2004-07-11 | | Release date: | 2004-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptidoglycan amidase MepA is a LAS metallopeptidase
J.Biol.Chem., 279, 2004
|
|
1U10
 
 | | MEPA, active form with ZN in P1 | | Descriptor: | Penicillin-insensitive murein endopeptidase, SULFATE ION, ZINC ION | | Authors: | Marcyjaniak, M, Odintsov, S.G, Sabala, I, Bochtler, M. | | Deposit date: | 2004-07-14 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptidoglycan amidase MepA is a LAS metallopeptidase
J.Biol.Chem., 279, 2004
|
|
5LEO
 
 | | Complex structure of lysostaphin SH3b domain with peptidoglycan fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLY-GLY-GLY-GLY-GLY, Lysostaphin, ... | | Authors: | Jagielska, E, Nowak, E, Bochtler, M, Sabala, I. | | Deposit date: | 2016-06-30 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complex structure of lysostaphin SH3doamin with peptidoglycan fragment
To Be Published
|
|
4ZYB
 
 | | High resolution structure of M23 peptidase LytM with substrate analogue | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Grabowska, M, Jagielska, E, Czapinska, H, Bochtler, M, Sabala, I. | | Deposit date: | 2015-05-21 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of an M23 peptidase with a substrate analogue.
Sci Rep, 5, 2015
|
|
7BNH
 
 | |
7BNG
 
 | |
7BNI
 
 | |
6SMK
 
 | | Crystal structure of catalytic domain A109H mutant of prophage-encoded M23 protein EnpA from Enterococcus faecalis. | | Descriptor: | Peptidase_M23 domain-containing protein, ZINC ION | | Authors: | Malecki, P.H, Mitkowski, P, Czapinska, H, Sabala, I. | | Deposit date: | 2019-08-22 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural Characterization of EnpA D,L-Endopeptidase from Enterococcus faecalis Prophage Provides Insights into Substrate Specificity of M23 Peptidases.
Int J Mol Sci, 22, 2021
|
|
2AYI
 
 | | Wild-type AmpT from Thermus thermophilus | | Descriptor: | Aminopeptidase T, ZINC ION | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-09-07 | | Release date: | 2005-11-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Substrate Access to the Active Sites in Aminopeptidase T, a Representative of a New Metallopeptidase Clan.
J.Mol.Biol., 354, 2005
|
|
1ZJC
 
 | | Aminopeptidase S from S. aureus | | Descriptor: | COBALT (II) ION, aminopeptidase ampS | | Authors: | Odintsov, S.G, Sabala, I, Bourenkov, G, Rybin, V, Bochtler, M. | | Deposit date: | 2005-04-28 | | Release date: | 2005-06-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Staphylococcus aureus Aminopeptidase S Is a Founding Member of a New Peptidase Clan.
J.Biol.Chem., 280, 2005
|
|
4OUN
 
 | | Crystal Structure of Mini-ribonuclease 3 from Bacillus subtilis | | Descriptor: | Mini-ribonuclease 3 | | Authors: | Chojnowski, G, Czarnecka, J, Nowak, E, Pianka, D, Glow, D, Sabala, I, Skowronek, K, Nowotny, M, Bujnicki, J.M. | | Deposit date: | 2014-02-18 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Sequence-specific cleavage of dsRNA by Mini-III RNase
Nucleic Acids Res., 43, 2015
|
|
1QWY
 
 | | Latent LytM at 1.3 A resolution | | Descriptor: | ZINC ION, peptidoglycan hydrolase | | Authors: | Odintsov, S.G, Sabala, I, Marcyjaniak, M, Bochtler, M. | | Deposit date: | 2003-09-03 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Latent LytM at 1.3A resolution.
J.Mol.Biol., 335, 2004
|
|
